Rhizosphere Bacterial Community and Diversity at Fields of Wilt Resistant or Susceptible Mulberry Trees
-
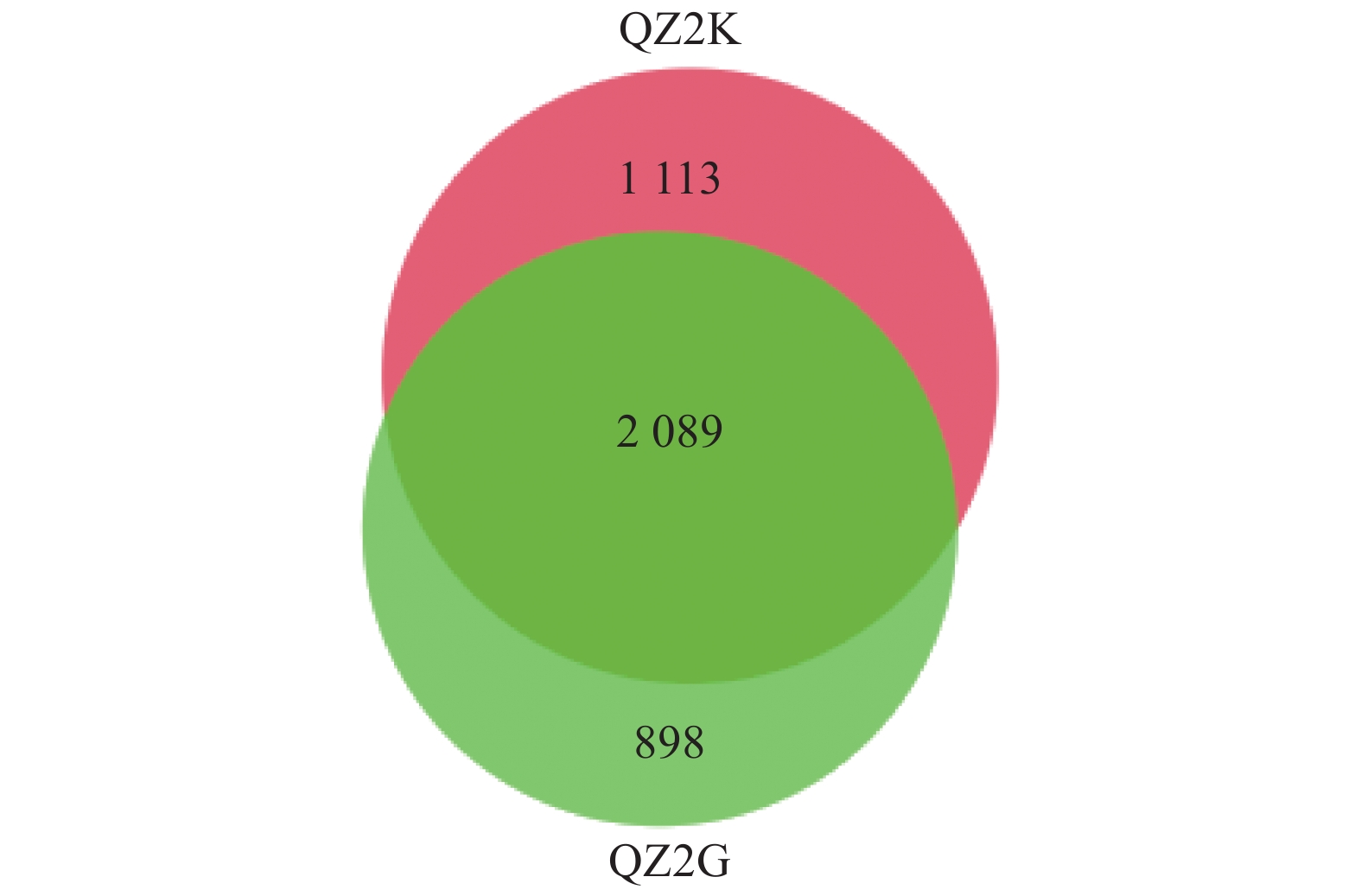
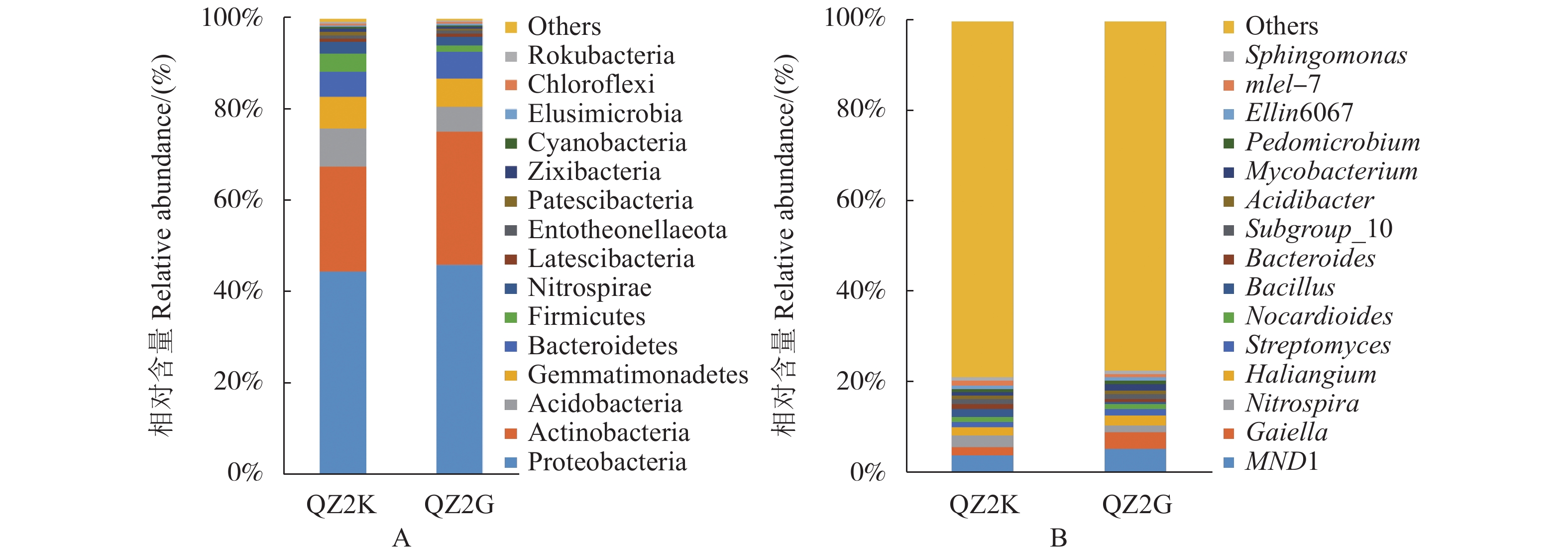
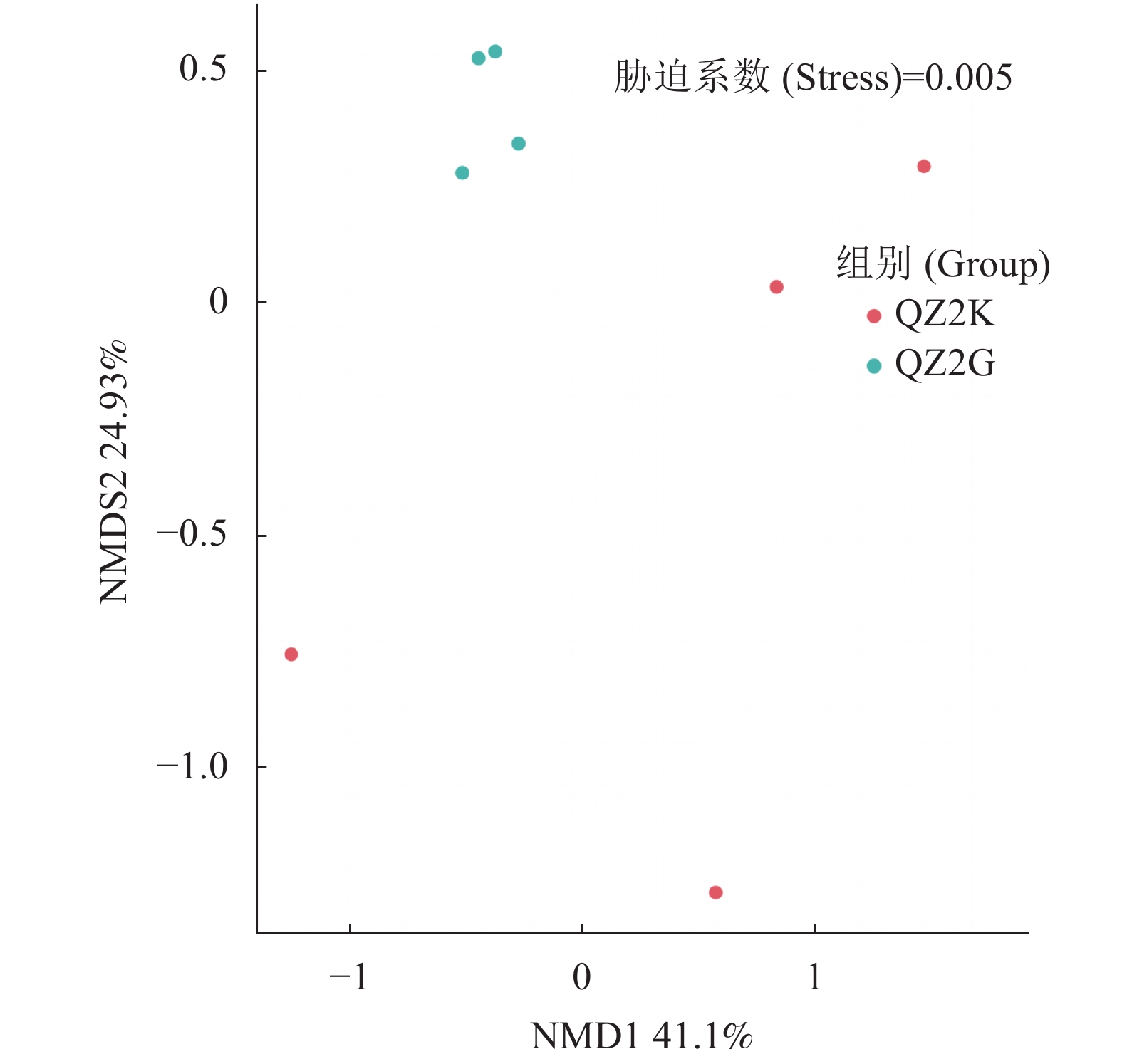
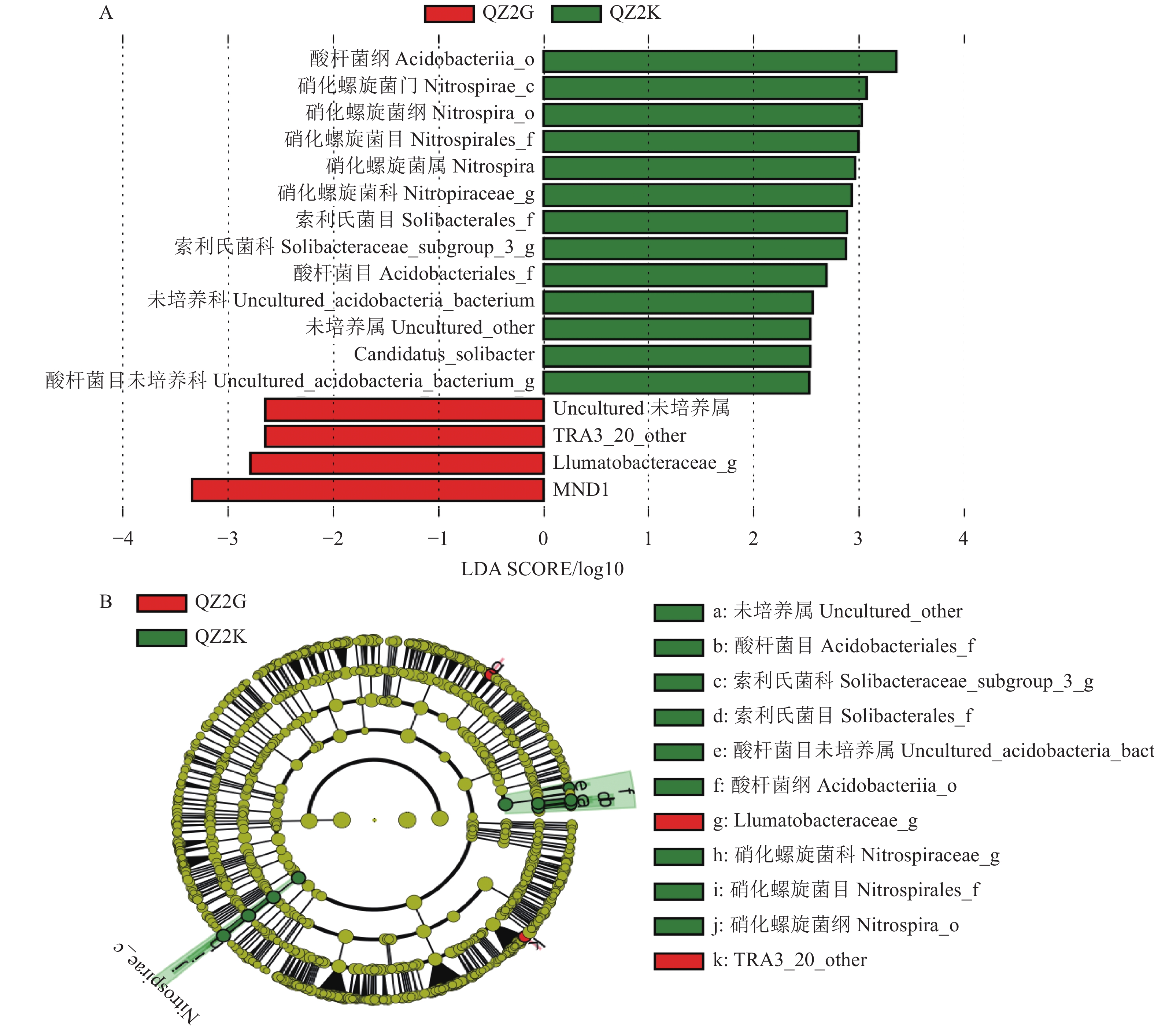
摘要:目的 通过抗、感青枯病桑树根际土壤细菌群落结构与多样性分析,阐明根际细菌群落与桑树抗、感青枯病的相关性。方法 采用Illumina MiSeq测序技术对抗青枯病桑树基因型(抗青283×抗青10)和感病基因型(桂桑优62)根际细菌的16S rRNA基因V3-V4区进行扩增,高通量测序后比较分析了其根际细菌群落结构与多样性。结果 ① 抗、感青枯病桑树根际土壤的优势菌门为变形菌门、放线菌门、酸杆菌门、芽单胞菌门和拟杆菌门,优势菌属为MND1、Gaiella、硝化螺菌属、Haliangium和链霉菌属;② 抗、感青枯病桑树根际土壤细菌群落Alpha多样性无显著差异,但经NMDS排序,则可显著区分(stress=0.005<0.05),其中抗性基因型根际土壤中具有重要作用的细菌类群较多,包括:硝化螺旋菌纲、酸杆菌纲、硝化螺旋菌目、索利氏菌目、酸杆菌目、硝化螺旋菌科、酸杆菌目未培养科、索利氏菌科Subgroup 3和Subgroup 2未培养科,而敏感基因型桂桑优62根际重要作用的细菌类群则主要为Ilumatobacteraceae和TRA3 20 Other两个科。③ 抗、感青枯病桑树根际细菌差异COG类目为763,仅占17.25%。结论 抗、感青枯病桑树根际土壤细菌群落丰富度和多样性虽无显著差异,但其重要作用的类群显著不同,说明桑树根际土壤细菌群落结构可能与种质对青枯病的抗性具有相关性。本结果将为进一步研究桑树根际微生态特征、筛选有益功能菌株并用于桑青枯病的有效防控具有重要意义。
-
关键词:
- 桑树 /
- 青枯病 /
- Illumina高通量测序 /
- 细菌多样性 /
- 群落结构
Abstract:Objective Structure and diversity of rhizosphere bacterial communities at fields of mulberry trees resistant (QZ2K) or susceptible (QZ2G) to wilt disease were studied.Method The V3-V4 regions of 16S rRNA in rhizosphere bacteria were amplified and sequenced using high-throughput sequencing technology on Illumina MiSeq to determine the bacterial community structure, diversity, and functions. Results from the two field samples were compared.Result (1) At phylum and genus levels, the dominant rhizosphere bacteria were similar at QZ2K (Kangqing 283×Kangqing 10 mulberry field) and at QZ2G (Guisangyou 62 mulberry field). The phyla included Proteobacteria, Actinobacteria, Acidobacteria, Gemmatimonadetes, and Bacteroidetes, while the genera consisted of MND1, Gaiella, Nitrospira, Haliangium, and Streptomyces. (2) Although no significant difference in the alpha diversity of the bacteria communities at the two different fields, the NMDS ordination showed significant differences (stress 0.005<0.05). At QZ2K, the bacteria related to significant metabolic functions were Nitrospira, Acidobacteriia, Nitrospirales, Solibacteraies, Acidobacteriales, Nitrospiraceae, and the uncultured Acidobacteria, Solibacteraies Subgroup 3, and Solibacteraies Subgroup 2. At QZ2G, only Ilumatobacteraceae and TRA3-20-other were identified. (3) According to the Wilcoxon signed rank test, the 763 different rhizosphere bacteria orthologs identified were only 17.25% of all COG orthologs on both fields.Conclusion There were no significant differences in the richness and diversity of rhizosphere bacteria community between the two fields. However, the bacteria associated with significant functions differed significantly between them, which could well be the species that made the difference in the occurrence of wilt disease on the mulberry plants. The information obtained in this study was of value for further studies on the microecological characteristics of mulberry rhizosphere as well as selection and application of functional bacteria for wilt control. -
0. 引言
【研究意义】油柰(Prunus salicina lindley)是蔷薇科(Rosaceae)李属(Prunus)核果类落叶果树,是福建省特色水果和稀优李资源,具有较高的经济价值[1],但逆境胁迫(如极端温度[2]、干旱[3]等)均会影响油柰的生长发育,严重时甚至造成减产[4]。植物对各种逆境胁迫的响应需通过一系列信号网络的感知、传导和传递[5],故研究植物在逆境胁迫下的信号感知传递过程及其分子机制,对研究植物抗逆机理,提高植物抗性具有重要意义。【前人研究进展】在植物应答各种非生物胁迫的过程中,作为植物体内最大转录因子家族之一的WRKY转录因子在调节植物应答逆境胁迫的防御反应中发挥着至关重要的作用[5-8]。研究表明,在拟南芥中,ABA、干旱、盐处理能诱导增强AtWRKY33
和AtWRKY25的表达,通过表达可增强拟南芥对盐的耐受性[9]。分别在拟南芥中过表达TaWRKY33[10]和CkWRKY33 [11]可提高转基因株系在高温胁迫和干旱胁迫下的存活率。在紫花苜蓿中过表达MsWRKY33 可以提高转基因材料的耐盐性[12]。在葡萄中VaERF092 通过结合VaWRKY33启动子GCC-box调控VaWRKY33,从而增强植物的耐寒性[13]。在油柰上,外源SA处理下能显著抑制PsWRKY22启动子在拟南芥中的表达,表明SA能调控PsWRKY22的表达[14]。【本研究切入点】基于前人相关研究发现有关油柰的抗性基因研究相对较少,本研究前期通过对油柰cDNA文库的筛选,获得一个能响应SA的WRKY基因并命名为PsWRKY33,经克隆PsWRKY33启动子全长以及缺失序列构建含有GUS报告基因的载体,通过转基因到拟南芥来分析PsWRKY33启动子不同片段在不同处理下的表达模式。【拟解决的关键问题】利用模拟各种非生物胁迫,探测转基因PsWRKY33启动子不同片段在不同胁迫处理下的应答模式,以期为进一步研究油柰WRKY基因生物学功能及其调控机制提供科学数据。 1. 材料与方法
1.1 植物材料和生长条件
以福建古田地区标准室外嫁接育苗的一年生翠屏晚柰品种作为研究对象。取叶片样品,迅速冷冻于液氮中用于提取油柰基因组DNA。
将野生型(Columbia)拟南芥以及转基因的种子播种于装有经高压灭菌后的基质中[V(泥炭苔):V(蛭石)=2∶1]。置于(25±1) ℃、光照强度为100 μmol·ms−1(12 h光 / 12 h暗循环)、相对湿度为60%的条件下培养。转基因拟南芥种子在4 ℃暗处理3 d后用灭菌水洗1次,后用75 %酒精洗3~5 min,最后用无菌水洗5次。转基因T0和T1代种子播种在含50 mg·L−1潮霉素的½MS固体培养基中进行筛选。将培养皿转移到培养箱中,在(23±1) ℃、相对湿度80%的条件(12 h 光/12 h暗循环)下培养。
1.2 生物信息学分析
在NCBI数据库blastp比对获得与PsWRKY氨基酸序列同源性较高的5种植物的蛋白序列,利用MEGA 6.06软件将以上6种植物氨基酸序列进行同源比对,构建系统进化树。
利用PlantCARE数据库(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)分析预测PsWRKY33
5′端上游启动子区域存在的顺式作用调控元件。 1.3 载体构建、植物转化、胁迫处理
利用染色体步移法从油柰基因组DNA中克隆出PsWRKY33
启动子片段,在此基础上利用PCR技术扩增出3个不同缺失片段(表1),载体构建过程、转基因过程、胁迫处理等过程参照陈永萍等[14]的方法。 表 1 引物序列Table 1. Sequences of primers引物名称 Primer name 引物序列(5′-3′) Sequence of primers(5′-3′) PsWRKY33-GSPA GCGAAGTAAGAAGAGGGAGAAACGGAG PsWRKY33-GSPB TGTTGGTGCTGGGGTCTTCATCTG PsWRKY33-GSPC CGAGAAAGTGAAGGTTGGGTGTGAA pPsWRKY33-F GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGTTCTGCCTTGTTTGCT pPsWRKY33-P1-F GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTGATCGAATTTTTTTTTCATA pPsWRKY33-P2-F GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATTATATTGGTTTACATACA pPsWRKY33-P3-F GGGGACAAGTTTGTACAAAAAAGCAGGCTTCAAACCCACGAGCTTTGACCA pPsWRKY33-R GGGGACCACTTTGTACAAGAAAGCTGGGTCAAAGGGGGGGAGAGAGAGAG 1.4 组织化学染色和荧光法检测GUS活性
采集不同胁迫处理下的7日龄T3转基因株系的叶片,以及T3代转基因株系的叶片、花序和角果进行GUS染色并拍照,具体操作方法过程参照陈永萍等[14]的方法。
参照陈永萍等[14]的方法,采用荧光法定量测定胁迫处理下的转基因植株GUS相对活性。数据整理后用SigmaPlot 12.5软件作图。所有试验重复3次。采用t检验进行统计学分析。
2. 结果与分析
2.1 PsWRKY33系统发育分析
为确定PsWRKY基因与其他物种之间的系统发育关系,将获得的PsWRKY氨基酸序列与梅花PmWRKY33(accession no. XP_008231402.2)、欧洲甜樱桃PaWRKY24L(accession no. XP_021810565.1)、东京樱花PyWRKY33(accession no. PQM41056.1)、巴旦木PpWRKY24(accession no. XP_007208290.1)和拟南芥AtWRKY33蛋白序列(accession no. NP_181381.2)进行系统进化树分析。结果表明,PsWRKY与拟南芥AtWRKY33具有较高的序列相似性。因此,我们将该基因命名为PsWRKY33
(图1)。 2.2 PsWRKY33启动子序列的克隆及顺式元件分析
利用染色体步移法获得PsWRKY33
的ATG起始密码子上游1 872 bp的片段,并利用PlantCARE网站对其启动子序列进行预测分析。结果发现(图2),PsWRKY33 启动子含有大量参与激素诱导响应、逆境胁迫响应等顺式作用元件,如分别响应低温、生长素、SA、ABA、乙烯等的LTR、AuxRR-core、TCA、ABRE、ERE等顺式元件。在此基础上,通过对不同的响应元件进行切除获得不同缺失片段,并分别构建到pMDC163表达载体上(图3)。经浸花法分别转化大野生型拟南芥中,经潮霉素筛选与PCR检测转基因阳性植株(图4),最终获得含有pPsWRKY33、P1、P2、P3的转基因株系,分别命名为pPsWRKY33::GUS、pPsWRKY33-P1::GUS、pPsWRKY33-P2::GUS和pPsWRKY33-P3::GUS。 ![]() 图 2 PsWRKY33
图 2 PsWRKY33基因启动子序列 注:-60表示ATG上游60个核苷酸。红色粗体字母加框表示预测的顺式作用元件序列。pPsWRKY33、P1、P2、P3加箭头加黑色下划线表示为4个片段(−1 872、−852、−357和−271 bp)的特异性引物,箭头表示它们的方向。Figure 2. Cis-elements of PsWRKY33 promoterNote: -60 represented the 60 nucleotides upstream of the ATG. Bold red letters with boxes indicated the predicted sequence of cis-acting elements. pPsWRKY33, P1, P2 and P3 add arrows and black underlines represented the specific primers of four fragments (−1 872, −852, −357 and −271 bp), the arrows indicated their direction.![]() 图 3 PsWRKY33
图 3 PsWRKY33启动子各片段的GUS载体 注:不同启动子区域的数目表示PsWRKY33翻译起始ATG密码子上游的核苷酸位置。attR1和attR2限制性位点:将pDMC163载体中的CaMV35S启动子替换为PsWRKY33启动子片段。LB和RB:代表T-DNA的左右边缘;Hyg:潮霉素标记基因;GUSA:GUS基因编码区域。黄色的条形图代表PsWRKY33::GUS、PsWRKY33-P1::GUS、PsWRKY33-P2::GUS和PsWRKY33-P3::GUS结构的-1 872 bp、-852 bp、-357 bp和-271 bp PsWRKY33启动子片段。Figure 3. Schematic diagram of GUS vector of individual PsWRKY33 promoter fragmentsNote: The number of different promoter regions represents the nucleotide position upstream of the PsWRKY33 translation initiation ATG codon. The CaMV35S promoter in pDMC163 vector was replaced by PsWRKY33 promoter fragment by attR1 and attR2 restriction sites. LB and RB represent the left and right edges of T-DNA; Hyg, hygromycin B marker gene; GUS is the region that codes for the GUS gene. The yellow bars represent the -1 872 bp, -852 bp, -357 bp and -271 bp PsWRKY33 promoter fragments of PsWRKY33::GUS, PsWRKY33-P1::GUS, PsWRKY33-P2::GUS and PsWRKY33-P3::GUS, respectively.![]() 图 4 PCR检测T2代转基因植株注:A~D分别为pPsWRKY33、pPsWRKY33-P1、pPsWRKY33-P2和pPsWRKY33-P3转基因植株的PCR检测结果。1~8分为pPsWRKY33、pPsWRKY33-P1、pPsWRKY33-P2和pPsWRKY33-P3 8个转基因植株;9为质粒;10为野生型;11为DL 2 000 DNA maker。Figure 4. Detectioned of transgenic Arabidopsis T2 plants by PCRNote: A-D: PCR analysis on pPsWRKY33, pPsWRKY33-P1, pPsWRKY33-P2, and pPsWRKY33-P3 transgenic Arabidopsis, respectively; 1-8: 8 strains of pPsWRKY33, pPsWRKY33-P1, pPsWRKY33-P2, and pPsWRKY33-P3 transgenic Arabidopsis, respectively; 9: plasmid; 10: wild type; 11: DNA maker DL 2 000.
图 4 PCR检测T2代转基因植株注:A~D分别为pPsWRKY33、pPsWRKY33-P1、pPsWRKY33-P2和pPsWRKY33-P3转基因植株的PCR检测结果。1~8分为pPsWRKY33、pPsWRKY33-P1、pPsWRKY33-P2和pPsWRKY33-P3 8个转基因植株;9为质粒;10为野生型;11为DL 2 000 DNA maker。Figure 4. Detectioned of transgenic Arabidopsis T2 plants by PCRNote: A-D: PCR analysis on pPsWRKY33, pPsWRKY33-P1, pPsWRKY33-P2, and pPsWRKY33-P3 transgenic Arabidopsis, respectively; 1-8: 8 strains of pPsWRKY33, pPsWRKY33-P1, pPsWRKY33-P2, and pPsWRKY33-P3 transgenic Arabidopsis, respectively; 9: plasmid; 10: wild type; 11: DNA maker DL 2 000.2.3 PsWRKY33启动子在不同组织的活性分析
利用GUS组织化学方法分析4个启动子片段在转基因拟南芥T3幼苗中的表达特征。结果表明,在转基因拟南芥T3代苗的叶片、花序和角果中,PsWRKY33启动子的4个缺失片段存在不同的表达模式。pPsWRKY33::GUS在叶片、花序和花序梗中均检测到较强的GUS活性,且随着启动子片段缩短,叶片和花序梗中的GUS活性越弱。4个缺失片段在角果上均未发现明显的GUS活性,其中在pPsWRKY33::GUS、pPsWRKY33-P1::GUS和pPsWRKY33-P2::GUS的角果荚底部观察到轻微的GUS活性(图5)。
2.4 4个PsWRKY33
启动子片段对不同胁迫的响应特征 为确定PsWRKY33启动子是否能响应不同的逆境胁迫,故对7日龄T3转基因拟南芥植株进行ABA、SA、MeJA、ACC(ETH)、NaCl、MAN和低温(4 ℃)处理12 h,检测其GUS染色(图6)以及相关GUS酶活(图7)。经GUS染色结果发现PsWRKY33启动子4个缺失片段在不同胁迫处理下的GUS活性强弱不同。不同PsWRKY33启动子片段在4 ℃低温处理下,各片段GUS活性均强于对照组,而在SA处理下4个缺失片段的GUS活性最弱,其中在全长、P1和P2缺失片段的植株发育真叶中检测到微弱的GUS活性,而在P3缺失片段的转基因植株中未能明显观察到GUS活性。同时发现4个PsWRKY33启动子片段转基因植株在对照组及NaCl和ABA处理下随着片段缺失长度的增加,其GUS活性逐渐减弱;而在MAN和ETH处理下,随着越长的片段缺失,其GUS活性反而明显增强。而在MeJA处理下,全长和P3缺失片段植株的GUS活性强于P1和P2缺失片段植物的GUS活性(图6)。
![]() 图 7 转基因拟南芥在不同胁迫处理下的GUS相对活性注:图中不同大、小写字母表示同一处理达极显著(P<0.01)、显著(P<0.05)差异。Figure 7. GUS relative activity in transgenic Arabidopsis under different stressesNote: Different uppercase and lowercase letters indicate highly significant (P<0.01) and significant (P<0.05) differences in the same treatment .
图 7 转基因拟南芥在不同胁迫处理下的GUS相对活性注:图中不同大、小写字母表示同一处理达极显著(P<0.01)、显著(P<0.05)差异。Figure 7. GUS relative activity in transgenic Arabidopsis under different stressesNote: Different uppercase and lowercase letters indicate highly significant (P<0.01) and significant (P<0.05) differences in the same treatment .为进一步定量分析各片段GUS活性在各胁迫下的变化趋势,同时对各胁迫下的4个PsWRKY33
启动子缺失片段转基因拟南芥幼苗进行GUS相对酶活测定分析。结果图7可见,不同长度的GUS活性在不同处理下呈现出不同的变化趋势。在对照组、4 ℃低温、盐、ABA、SA处理下,各片段GUS活性表现出片段越短,其GUS活性越低的变化模式,并且发现在除了乙烯处理外的各个处理下,全长GUS活性均不同程度上高于3个缺失片段。在乙烯和MeJA处理下,全长和P3的GUS活性高于P1和P2的GUS活性。相比于对照组,4 ℃低温处理下各片段GUS活性表现出不同程度上地提高,而ABA和SA处理下,4个片段GUS活性显著降低。总体上GUS活性变化趋势与GUS染色结果相似。 综上所述,在对照组及4 ℃、盐、ABA和SA的胁迫下,随着缺失的片段越长,其GUS活性下降越明显。这表明,PsWRKY33
启动子的3个缺失片段失去对4 ℃、ABA、SA和NaCl胁迫响应的特性。 3. 讨论与结论
WRKY转录因子是植物体内最大的转录因子家族之一,广泛参与植物生长发育过程,以及对各逆境胁迫(生物、非生物及激素胁迫)的响应[15]。WRKY转录因子通过激活相关信号通路[16],与各种胁迫应答基因启动子区的顺式作用元件相互作用,正向或负向调节激素相关基因或各种胁迫防御应答基因的表达,参与防御反应,促使植物在分子、生理生化等水平做出调节以适应各种环境胁迫[17]。启动子是基因转录起始位点前的一段重要序列,对基因的转录调控起着重要影响[18]。油柰上发现PsWRKY22基因能被外源激素SA处理诱导差异表达[19],在拟南芥中过表达PsWRKY22基因启动子[14]发现该基因启动子在SA处理下表达被显著抑制,为研究同样能被外源激素SA处理诱导差异表达的PsWRKY33基因[19]转录调控机制,本研究借鉴PsWRKY22基因启动子[14]应答不同胁迫处理的研究方法,以期探讨PsWRKY33启动子的功能和表达调控的分子机制。
启动子序列上包含核心元件及一些响应逆境胁迫的顺式元件,研究顺式作用元件对启动子以及基因功能的作用,对分析目的基因的功能具有重要意义[18]。PsWRKY22基因启动子上含有响应多个响应激素、逆境相关的顺式元件[14],尤其含有SA响应元件TCA-element,经试验发现PsWRKY22基因启动子在SA处理下活性被抑制表达与该元件有关。本研究经网页预测也发现PsWRKY33基因启动子区域含有多种响应激素、逆境相关的顺式作用元件,如ABA响应元件ABRE、生长素响应元件AuxRR-core、ETH响应元件ERE、GA响应元件P-box和TATC-box、SA响应元件TCA、低温响应元件LTR、响应干旱和ABA元件MYB、响应干旱、ABA和低温胁迫元件MYC、响应机械损伤元件WUN-motif,推测该基因的表达可能会受ABA、ETH、低温等的诱导或抑制。
高永峰等[20]分析显示SlWRKY31基因启动子上含有分别响应热胁迫、干旱、ABA和SA的HSE、MBS、ABRE和TCA-element元件,通过不同胁迫处理后发现该启动子显著受到盐、甘露醇、SA、ABA和42 ℃高温的诱导表达,说明SlWRKY31基因启动子是一个可以响应多种逆境胁迫的诱导型启动子[14]。刘志钦等[21]通过构建CaWRKYK5启动子5′端不同缺失片段的载体,经青枯病菌、激素等处理分析后得出CaWRKYK5启动子应答青枯菌、MeJA等的顺式元件区域。本研究经多种胁迫处理,结果发现在SA处理下,PsWRKY33启动子GUS染色和活性均显著低于对照组,说明PsWRKY33可能参与油柰对过量SA胁迫的响应,进而通过调节SA去响应逆境胁迫。而在极端温度(4 ℃低温)胁迫下,GUS染色和活性均高于对照,说明该基因可能是受低温诱导的。虽然在本研究没有预测到应答JA的顺式作用元件,但是在JA的处理下,PsWRKY33启动子驱动的GUS报告基因可以被激活,说明可能存在其他的未知的调控机制。
PsWRKY33基因启动子能被外源ABA、SA抑制转录表达活性,而受到低温诱导表达,可能与其含有的ABRE、LTR和TCA元件有关。在ETH处理后,pPsWRKY33和pPsWRKY33-P1的GUS活性受到抑制,而pPsWRKY33-P2/P3的GUS活性被诱导上调表达,推测−1 871 ~ −852 bp区域可能含有对ETH应答起着负调控的顺式元件,而−357 bp ~ −1 (ATG)区域里可能存在应答ETH的核心元件。
本研究检测不同胁迫处理下PsWRKY33基因启动子及其缺失片段的GUS活性,但还未确定具体发挥作用的顺式作用元件,以及对下游基因表达起促进或抑制作用的是由于单个顺式作用元件调控引起的,还是由多个顺式元件协同作用引起的结果[22],已有文献表明,通过对启动子上的元件进行缺失处理分析的方法,是作为鉴定启动子顺式作用元件功能的重要手段之一[23]。Rushton等[24]通过对PR1基因启动子进行缺失及功能分析后发现,对启动子W-box元件核心序列进行突变后启动子会丧失响应病原菌诱导的能力,故日后可通过对PsWRKY33基因启动子上的顺式元件碱基进行点突变或其他方法来确定其启动子应答各逆境胁迫的核心元件位置,为后续对PsWRKY33基因及其启动子的功能和表达调控的分子机制研究奠定坚实的基础。
-
图 5 抗感青枯病桑树根际细菌预测得到的COG相对丰度
注:A: RNA加工与修饰; B: 染色体结构与动态;C: 能量产生与转化; D: 细胞周期控制,细胞分裂,染色体分裂; E: 氨基酸转运与代谢; F: 核酸转运与代谢;G:碳水化合物的运输与代谢; H: 酶运输与代谢; I: 脂质运输与代谢; J:翻译,核糖体结构和生物发生; K: 转录; L: 复制,重组和修复; M: 细胞壁/膜/包膜生物发生; N: 细胞运动; O: 翻译后修饰,蛋白质更新,伴侣; P: 无机离子的运输与代谢; Q:次生代谢产物的生物合成,转运和分解代谢; R:仅通用功能预测; S:未知功能; T:信号转导机制; U:细胞内运输,分泌和囊泡运输; V: 防御机制; W: 细胞外结构; Z: 细胞骨架。
Figure 5. Relative abundance of COG at QZ2K and QZ2G
Note: A: RNA processing and modification; B: Chromatin structure and dynamics; C: Energy production and conversion; D: Cell cycle control, cell division, and chromosome partitioning; E: Amino acid transport and metabolism; F: Nucleotide transport and metabolism; G: Carbohydrate transport and metabolism; H: Coenzyme transport and metabolism; I: Lipid transport and metabolism; J: Translation, ribosomal structure and biogenesis; K: Transcription; L: Replication, recombination, and repair; M: Cell wall/membrane/envelope biogenesis; N: Cell motility; O: Posttranslational modification, protein turnover, and chaperones; P: Inorganic ion transport and metabolism; Q: Secondary metabolites biosynthesis, transport, and catabolism; R: General function prediction only; S: Function unknown; T: Signal transduction mechanisms; U: Intracellular trafficking, secretion, and vesicular transport; V: Defense mechanisms; W: Extracellular structures; Z: Cytoskeleton.
表 1 高通量测序数据概况
Table 1 Overview of high-throughput sequencing data
样品
Sample高质量序列
Clean tags优质序列
Valid tags平均长度 /bp
Valid mean length测序深度指数/%
Goods coverageQZ2K.1 33 883 28 948 429 94.81 QZ2K.2 38 786 32 371 430 94.69 QZ2K.3 32 328 26 941 432 94.57 QZ2K.4 41 499 34 267 432 94.56 平均值 average 36 624.0 30 631.8 430.8 94.658 QZ2G.1 38 747 32 496 432 94.53 QZ2G.2 37 626 31 688 432 94.43 QZ2G.3 40 749 34 814 432 94.78 QZ2G.4 40 656 34 090 432 95.22 平均值 average 3 944.5 33 272.0 432.0 94.740 表 2 抗感青枯病桑树根际土壤细菌α多样性指数(平均值±标准误,n=4)
Table 2 Alpha diversity indices of bacteria in rhizosphere soils at QZ2K and QZ2G(Mean±SE,n=4)
样品
Samples谱系多样性指数
PD whole TreeChao1 文库覆盖率(%)
Goods coverage观测物种数
Observed Species香浓-威纳指数
Shannon-Wiener辛普森指数
SimpsonQZ2K 84.97±0.77 3 190.57±22.48 0.95±0.00 2 333.88±40.49 9.51±0.09 0.995±0.001 QZ2G 82.86±1.44 3 178.61±87.45 0.95±0.00 2 304.30±49.58 9.65±0.03 0.997±0.001 -
[1] XIONG W, SONG Y Q, YANG K M, et al. Rhizosphere protists are key determinants of plant health [J]. Microbiome, 2020, 8: 27. DOI: 10.1186/s40168-020-00799-9
[2] RODRIGUEZ P A, ROTHBALLER M, CHOWDHURY S P, et al. Systems biology of plant-microbiome interactions [J]. Molecular Plant, 2019, 12(6): 804−821. DOI: 10.1016/j.molp.2019.05.006
[3] BERENDSEN R L, VISMANS G, YU K, et al. Disease-induced assemblage of a plant-beneficial bacterial consortium [J]. The ISME Journal, 2018, 12(6): 1496−1507. DOI: 10.1038/s41396-018-0093-1
[4] ROUT M E, SOUTHWORTH D. The root microbiome influences scales from molecules to ecosystems: The unseen majority1 [J]. American Journal of Botany, 2013, 100(9): 1689−1691. DOI: 10.3732/ajb.1300291
[5] KWAK M J, KONG H G, CHOI K, et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato [J]. Nature Biotechnology, 2018, 36(11): 1100−1109. DOI: 10.1038/nbt.4232
[6] ROLFE S A, GRIFFITHS J, TON J. Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes [J]. Current Opinion in Microbiology, 2019, 49: 73−82. DOI: 10.1016/j.mib.2019.10.003
[7] WEI Z, GU Y A, FRIMAN V P, et al. Initial soil microbiome composition and functioning predetermine future plant health [J]. Science Advances, 2019, 5(9): eaaw0759. DOI: 10.1126/sciadv.aaw0759
[8] 颜朗, 张义正, 方志荣, 等. 不同马铃薯基因型对根际细菌群落结构的影响 [J]. 四川大学学报(自然科学版), 2020, 57(2):383−390. YAN L, ZHANG Y Z, FANG Z R, et al. Effects of potato genotype on rhizososphere bacterial community structure [J]. Journal of Sichuan University (Natural Science Edition), 2020, 57(2): 383−390.(in Chinese
[9] LAREEN A, BURTON F, SCHÄFER P. Plant root-microbe communication in shaping root microbiomes [J]. Plant Molecular Biology, 2016, 90(6): 575−587. DOI: 10.1007/s11103-015-0417-8
[10] 蔡秋华, 左进香, 李忠环, 等. 抗性烤烟品种根际微生物数量及功能多样性差异 [J]. 应用生态学报, 2015, 26(12):3766−3772. CAI Q H, ZUO J, LI Z H, et al. Difference of rhizosphere microbe quantity and functional diversity among three flue-cured tobacco cultivars with different resistance [J]. Chinese Journal of Applied Ecology, 2015, 26(12): 3766−3772.(in Chinese
[11] MENDES L W, RAAIJMAKERS J M, DE HOLLANDER M, et al. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function [J]. The ISME Journal, 2018, 12(1): 212−224. DOI: 10.1038/ismej.2017.158
[12] 曹梦琪. 桑树青枯病病原菌的分离鉴定及其活体检测方法的建立 [D]. 镇江: 江苏科技大学, 2017. CAO M Q. Isolation, Identification and Establishment of Viable Cells Detection of the Pathogen of Mulberry Bacterial Wilt [D]. Zhenjiang: Jiangsu University of Science and Technology, 2017. (in Chinese).
[13] 李磊. 华南地区桑树青枯病原菌的收集鉴定及侵染机制的初步研究 [D]. 镇江: 江苏科技大学, 2017. LI L. Collecting for identification of the pathogen of Mulberry Wilt Disease in South China and a preliminary study on the mechanism of infection [D]. Guangzhou: Jiangsu University of Science and Technology, 2017. (in Chinese).
[14] 王树昌, 耿涛, 黄华平. 海南蚕桑 [M]. 海口: 南海出版公司, 2017: 68-80. [15] 黄富, 张伟国, 黄胜, 等. 桑树抗青283×抗青10杂交组合的育成 [J]. 广东蚕业, 2006, 4(4):24−28. HUANG F, ZHANG WG, HUANG S, et al. Breeding of mulberry Kangqing 283 × Kangqing 10 [J]. Guangdong Sericulture, 2006, 4(4): 24−28.(in Chinese
[16] 朱方荣. 桂桑优62和桂桑优12的特点及栽培 [J]. 广西农业科学, 2001(4):199. ZHU F R. Characteristics and cultivation of Guisangyou 62 and Guisangyou 12 [J]. Guangxi Agricultural Sciences, 2001(4): 199.(in Chinese
[17] NOSSA C W. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome [J]. World Journal of Gastroenterology, 2010, 16(33): 4135. DOI: 10.3748/wjg.v16.i33.4135
[18] 邱洁, 徐丽丽, 钱叶, 等. 不同品种桑树根际土壤细菌群落及土壤理化性质的研究 [J]. 蚕业科学, 2017, 43(4):568−576. QIU J, XU L L, QIAN Y, et al. An investigation on rhizospheric bacterial community and soil physical and chemical properties of different mulberry varieties [J]. Acta Sericologica Sinica, 2017, 43(4): 568−576.(in Chinese
[19] BHATTI A A, HAQ S, BHAT R A. Actinomycetes benefaction role in soil and plant health [J]. Microbial Pathogenesis, 2017, 111: 458−467. DOI: 10.1016/j.micpath.2017.09.036
[20] 向立刚, 周浩, 汪汉成. 健康与感染青枯病烟株根际土壤与茎秆细菌群落结构与多样性 [J]. 微生物学报, 2019, 59(10):1984−1999. XIANG LG, ZHOU H, WANG H C. Bacterial community structure and diversity of rhizosphere soil and stem of healthy and bacterial wilt tobacco plants [J]. Acta Microbiologica Sinica, 2019, 59(10): 1984−1999.(in Chinese
[21] KYSELKOVÁ M, KOPECKÝ J, FRAPOLLI M, et al. Comparison of rhizobacterial community composition in soil suppressive or conducive to tobacco black root rot disease [J]. The ISME Journal, 2009, 3(10): 1127−1138. DOI: 10.1038/ismej.2009.61
[22] ADHIKARI T B, JOSEPH C M, YANG G P, et al. Evaluation of bacteria isolated from rice for plant growth promotion and biological control of seedling disease of rice [J]. Canadian Journal of Microbiology, 2001, 47(10): 916−924. DOI: 10.1139/w01-097
[23] HUANG A C, JIANG T, LIU Y X, et al. A specialized metabolic network selectively modulates Arabidopsis root microbiota [J]. Science, 2019, 364(6440): 1−9.
[24] COTTON T E A, PÉTRIACQ P, CAMERON D D, et al. Metabolic regulation of the maize rhizobiome by benzoxazinoids [J]. The ISME Journal, 2019, 13(7): 1647−1658. DOI: 10.1038/s41396-019-0375-2
[25] BULGARELLI D, ROTT M, SCHLAEPPI K, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota [J]. Nature, 2012, 488(7409): 91−95. DOI: 10.1038/nature11336
[26] VÍCTOR J C, JUAN P J, VIVIANE C, et al. Pathogen-induced activation of disease - suppressive functions in the endophytic root microbiome [J]. Science, 2019, 366(6465): 606−612. DOI: 10.1126/science.aaw9285
-
期刊类型引用(1)
1. 邓江霞,张国良,李边豪,黄志炜,赵宏亮,张叶. WRKY类转录因子在植物激素ABA、SA和JA信号转导中的作用. 黑龙江农业科学. 2023(10): 138-144 .  百度学术
百度学术
其他类型引用(0)




 下载:
下载: