A Preliminary Study on Genetic and Serological Clustering of Pathogenic Pseudomonas plecoglossicida in Larimichthys crocea
-
摘要:目的 内脏白点病是冬春季大黄鱼(Larimichthys crocea)最主要的细菌性疾病,变形假单胞菌(Pseudomonas plecoglossicida)是内脏白点病的主要病原。分析大黄鱼来源的变形假单胞菌病原聚类特征,为大黄鱼变形假单胞菌流行规律研究和科学防控提供参考。方法 克隆7株不同时空来源的大黄鱼变形假单胞菌16S rDNA、gyrB,测序并构建进化树;制备上述菌株的O-特异链血清(O抗原血清)与鞭毛蛋白血清(H抗原血清),进行菌体凝集试验。结果 16S rDNA和gyrB序列分析表明,所有的变形假单胞菌聚为一簇;O抗原和H抗原凝集结果表明,7株变形假单胞菌以及变形假单胞菌标准株均会发生凝集反应,凝集效价也无明显区别,与荧光假单胞菌(Pseudomonas fluoresenes)、溶藻弧菌(Vibrio alginolyticus)和哈维氏弧菌(Vibrio harveyi)不发生凝集反应。结论 本研究所用的7株不同时空来源的变形假单胞菌亲缘关系接近,可能具有共同的起源。Abstract:Objective Clustering of strains of Pseudomonas plecoglossicida, a major pathogen that causes the serious bacterial visceral white spot disease on Larimichthys crocea in winter and spring, was studied for disease control and epidemic prevention.Method The 16S rDNA and gyrB of 7 suspected P. plecoglossicida strains collected from different temporal and spatial sources were cloned and sequenced with evolutionary trees constructed. O and H antigen serums of these strains were prepared for agglutination tests.Result The 7 pathogens could be clustered into a same branch based on their sequences. Their O and H antigen sera agglutinated with those of the standard P. plecoglossicida reference without significant differences on titers, but not with that of P. fluoresenes, Vibrio alginolyticus, or V. harveyi.Conclusion It appeared that the 7 collected strains of P. plecoglossicida were genetically closely related and might have a common origin.
-
0. 引言
【研究意义】二氧化硫是葡萄酒等果酒酿造中常用的添加剂,具有防止酒体过度氧化、挥发酸含量上升和抑制杂菌生长等作用[1]。但高量的二氧化硫(60 mg·L−1以上)会对酒体中的乳酸菌产生胁迫作用,使其生长受抑制甚至死亡,影响了果酒的苹果酸-乳酸发酵(Malolactic fementation,MLF)生物降酸效果和酒体品质提升[2];而低量的二氧化硫又不能充分发挥作用,从而易导致酒体品质劣变;因此高耐受二氧化硫的MLF乳酸菌的选择备受关注。植物乳杆菌(Lactobacillus plantarum)R23是本试验人员从自然发酵的枇杷酒中分离出的高耐硫、高抗逆性优良MLF乳酸菌,已在山葡萄、枇杷和杨梅等高酸性水果的酿造中广泛应用,成为果酒生物降酸中的重要菌株[3-4]。【前人研究进展】已有研究表明,二氧化硫在液体介质中以HSO3−、SO32−等形式存在,这些离子转化为SO42−的过程会产生活性氧簇(Reactive oxygen species,ROS)如∙O2ˉ、H2O2、∙OH等;持续增加的ROS将作用于细胞内生物大分子进而诱发核酸、蛋白质和脂质等的氧化损伤[5-6]。但同时,生物体会启动氧化防御系统以清除过多的ROS进而维持其正常的生长代谢,即所谓的氧化应激。事实上,强氧化环境均会导致生物体内ROS大量积累,引发生物体氧化应激反应[7],如槲皮素、芦丁等诱发了嗜酸乳杆菌NCFM氧化应激,且主要依靠过氧化氢酶的作用清除胞内自由基[8];另外,在李斯特氏菌的氧化应激反应中,超氧化物歧化酶活性和DNA损伤修复主导了自由基的清除过程[9]。【本研究切入点】推测二氧化硫胁迫可能引发植物乳杆菌R23氧化应激反应,且其抗氧化相关系统发挥了积极作用,但迄今植物乳杆菌R23耐受二氧化硫的应激机制尚不明确。【拟解决的关键问题】本研究从胞内抗氧化酶活力、细胞形态、细胞膜丙二醛和磷脂脂肪酸含量等方面进行探究,明确二氧化硫胁迫下植物乳杆菌R23生理代谢变化,以期揭示植物乳杆菌R23对二氧化硫胁迫的适应性机制。
1. 材料与方法
1.1 试验材料
1.1.1 菌株
植物乳杆菌(Lactobacillus plantarum)R23,由本实验室人员从自然发酵的枇杷酒中分离并保存,NCBI号为HQ58056。
1.1.2 培养基
LHR20液体培养基:酵母膏0.74%,牛肉膏1.0%,蛋白胨0.5%,蔗糖2.0%,柠檬酸铵0.2%,MgSO4 0.036%,MnSO4 0.005%,吐温-80 0.1%,苹果酸钠5.7%(体积分数35%),番茄汁5%,平菇浸汁5%,0.2 mol·L−1 Na2HPO4-0.2 mol·L−1 KH2PO4 缓冲盐稀释4倍,121 ℃湿热灭菌20 min。
1.1.3 主要仪器
SCIENTZ-950E细胞破碎仪(宁波新芝生物科技股份有限公司),SPX-250BS-Ⅱ型生化培养箱(上海新苗医疗器械制造有限公司),SW-CJ-IFD型单人单面净化工作台(苏州净化设备有限公司),TGL-18C型高速冷冻离心机(上海安亭科学仪器厂),GI54DW型高压灭菌器(美国致微),6380LV型扫描电镜(日本电子公司),BioTek Epoch2全波长酶标仪(Bio Tek), UV-1705紫外可见分光光度计(日本岛津),Agilent7890N型气相色谱系统(美国MIDI公司)。
1.1.4 主要试剂
(1)抗氧化酶活力测定及丙二醛含量测定试剂盒(上海起子生物科技有限公司)。(2)磷脂脂肪酸提取试剂。皂化试剂(试剂1):NaOH 45 g + 甲醇(HPLC grade)150 mL+去离子蒸馏水150 mL和甲醇混合后加入NaOH中,同时搅拌至完全溶解;甲基化试剂(试剂2):6.0 mol·L−1盐酸 325 mL+甲醇(HPLC grade)275 mL把盐酸加入到甲醇中,并不断搅拌;萃取试剂(试剂3):正己烷 (HPLC Grade ) 200 mL+MTBE (HPLC Grade ) 200 mL,把MTBE加入到正己烷中,并搅拌均匀;洗涤试剂(试剂4):NaOH 10.8 g+去离子蒸馏水900 mL。
1.2 试验方法
1.2.1 菌种活化
将植物乳杆菌R23甘油冻存种以5%(V/V)接种于LHR20中,30 ℃培养15~18 h,连续转接活化2次,备用。
1.2.2 菌体二氧化硫胁迫培养
将植物乳杆菌R23活化种以体积分数为5%接种于二氧化硫质量浓度分别为0、40、80、120 mg·L−1的 LHR20液体培养基中,30 ℃恒温静置培养4 h后,备用。
1.2.3 菌体处理与形态观察
取1 mL不同质量浓度二氧化硫胁迫处理后的菌液于1.5 mL无菌离心管中,5000 r·min−1离心3 min,用无菌水洗涤2次后,用适量2.5%戊二醛(pH 7.0)固定,后续制样、扫描电镜观察均在福建省农业科学院农业质量标准与检测技术研究所进行。
1.2.4 抗氧化酶活力测定样品制备
取50 mL不同质量浓度二氧化硫胁迫处理的植物乳杆菌R23菌悬液,以9000 r·min−1、4 ℃离心10 min,弃上清;加无菌生理盐水洗涤1次;菌泥中加入5 mL PBS(pH8.0)缓冲液,加玻璃珠旋涡震荡混匀;然后进行超声破碎,条件为:功率400 W、工作总时间5 min、工作1 s、间歇2 s、变幅杆直径5 mm;细胞破碎液经12000 r·min−1、4 ℃离心10 min;将上清液装于10 mL离心管,4 ℃暂存,备用。
1.2.5 蛋白质含量测定
采用考马斯亮蓝法。标准曲线的制作:取6支具塞试管,编号,按表1加入试剂,摇匀,反应10 min,于595 nm处测定吸光度。
表 1 蛋白质含量测定标准曲线Table 1. Standard curve for protein determination试剂
Reagent管号
Tube No.1 2 3 4 5 6 蛋白质标准液
Standard solution of protein/mL0 0.2 0.4 0.6 0.8 1.0 蒸馏水
Distilled water/mL1.0 0.8 0.6 0.4 0.2 0 考马斯亮蓝G-250试剂
Coomassie brilliant blue G-250 reagent/mL5 5 5 5 5 5 样品测定:取样品溶液0.1 mL于试管中,加水0.9 mL,各加入5 mL考马斯亮蓝试剂,充分混合,放置10 min,以试剂空白为对照,595 nm处测定吸光值,同时做3个重复。
1.2.6 酶活力及丙二醛含量测定
超氧化物歧化酶SOD、谷胱甘肽过氧化物酶GPX、过氧化氢酶CAT活力及丙二醛MDA含量的测定,采用试剂盒法,具体操作按照说明书进行。
1.2.7 细胞膜脂肪酸提取
(1)获菌:取植物乳杆菌R23胁迫前、后(分别用0、80 mg·L−1二氧化硫处理)的菌液20 mL,4 ℃、5000 r·min−1,离心5 min,用接种环挑取约40 mg湿重的菌落置于清洁干燥的有螺旋盖的试管(13 mm × 100 mm)底部;(2)皂化:在装有菌体的试管内加入(1.0±0.1)mL试剂1,锁紧盖子,震荡试管5~10 s,95~100 ℃水浴5 min,从沸水中移开试管并轻微冷却,震荡5~10 s,再水浴25 min,取出室温冷却;(3)甲基化:加入(2.0±0.1)mL试剂2,拧紧盖子,震荡5~10 s,80 ℃水浴10 min,移开且快速用流动自来水冷却至室温;(4)萃取:加入(1.25±0.1)mL 试剂3萃取溶剂,盖紧盖子,温和混合旋转10 min ,打开管盖,利用干净的移液管取出下层似水部分,弃去;(5)基本洗涤:加入(3.0±0.21)mL试剂4,拧紧盖子,温和混合旋转5 min,打开盖子,利用干净的移液管移出约2/3体积的上层有机相到干净的GC检体小瓶。
1.2.8 脂肪酸检测
应用MIDI系统,色谱分析柱温采用二阶顺序升温法,即第一阶段170 ℃起始,每分钟升温5 ℃,升至260 ℃,第二阶段每分钟升温40 ℃,升至310 ℃,维持90 s;汽化室温度250 ℃、检测器温度300 ℃;载气为氢气(2 mL·min−1)、尾吹气为氮气(30 mL∙min−1);柱前压68.95 kPa;进样量1 μL,进样分流比100∶1。将检测出的脂肪酸数据转换成以乳酸菌为样本、以脂肪酸生物标记为指标的数据矩阵Excel文件,然后利用生物统计软件SPSS 23.0中的Graphs进行分析。
2. 结果与分析
2.1 二氧化硫胁迫激发的菌体抗氧化防御反应
植物乳杆菌R23在梯度二氧化硫胁迫处理条件下的胞内抗氧化酶活力、膜丙二醛含量和超微形态变化如图1、图2和图3所示。以正常培养条件为对照,二氧化硫胁迫处理后3种抗氧化酶活力均显著提升(P < 0.05),其中80 mg·L−1 二氧化硫胁迫下SOD、CAT、GPX分别是无胁迫处理的1.64、2.14、1.62倍,特别是CAT活力提升速率最快且后续下降幅度最小,提示适量二氧化硫胁迫可有效激发植物乳杆菌R23的抗氧化系统,而CAT可能起了主导作用。当胁迫二氧化硫质量浓度增大到120 mg·L−1时,酶活力略有下降但与最高值无显著差异,且显著高于对照值(P<0.05)(图1)。
![]() 图 1 梯度二氧化硫胁迫下植物乳杆菌R23胞内抗氧化酶活力小写字母表示梯度二氧化硫质量浓度胁迫后菌体抗氧化酶活性差异显著(P<0.05)。Figure 1. Intracellular antioxidase activity of L. plantarum R23 under gradient of SO2 stressData with different lowercase letters represent significant differences on antioxidase activity of L. plantarum R23 treated by gradient of SO2 at P<0.05
图 1 梯度二氧化硫胁迫下植物乳杆菌R23胞内抗氧化酶活力小写字母表示梯度二氧化硫质量浓度胁迫后菌体抗氧化酶活性差异显著(P<0.05)。Figure 1. Intracellular antioxidase activity of L. plantarum R23 under gradient of SO2 stressData with different lowercase letters represent significant differences on antioxidase activity of L. plantarum R23 treated by gradient of SO2 at P<0.05图2显示,梯度二氧化硫胁迫导致MDA含量不断增加;其中40或80 mg·L−1二氧化硫胁迫下MDA变化幅度较小,之后加速上升 (P <0.05),在120 mg·L−1 二氧化硫胁迫下MDA含量是对照处理的1.84倍;说明抗氧化酶活力下降后胞内ROS不能得到有效清除,进而导致菌体脂质过氧化反应加剧。
植物乳杆菌R23在不同质量浓度二氧化硫胁迫下的菌体细胞超微形态如图3所示。正常培养条件下(0 mg·L−1二氧化硫),菌体饱满,呈杆状、单个或短链状排列,不生孢(图3-a);40 mg·L−1二氧化硫胁迫并未导致菌体形态发生肉眼看见的变化(图3-b);但胁迫程度进一步加深后,有些菌体表面发生皱缩现象,细胞失去原有的饱满程度(图3-c、d),其中120 mg·L−1二氧化硫胁迫下这一变化更加明显。可见80 mg·L−1 SO2胁迫下菌体尚可维持基本的形态,而120 mg·L−1二氧化硫胁迫虽未导致抗氧化酶活力显著下降,但其防御性能下降,这不仅表现在MDA含量显著提升,也表现在菌体形态的变化上。
2.2 二氧化硫胁迫诱导的细胞膜磷脂脂肪酸变化
上述研究结果表明,80 mg·L−1二氧化硫胁迫可有效激发植物乳杆菌R23生理应答,且未导致明显的结构损伤,因此考察了该浓度胁迫下菌体细胞膜磷脂脂肪酸的结构变化,结果如表2和图4所示。无胁迫时菌体细胞膜脂肪酸的组成成分包括饱和脂肪酸即十四烷酸(C14:0)、十六烷酸(C16:0)、十八烷酸(C18:0)、十二碳异脂肪酸(iso-C12:0)、十七碳前异脂肪酸(anteiso-C17:0),单不饱和脂肪酸即十六碳异脂肪酸(iso-C16:1)、十六碳顺式脂肪酸(iso-C16:1)和十九碳反式脂肪酸(trans-C19:1),以及环丙烷脂肪酸(cyclo-19:0)。其中C14:0、C16:0、C18:0和cyclo-19:0含量之和超过57.2%,占总脂肪酸的比例较大,特别是C16:0占总量的27%以上,应为植物乳杆菌R23脂肪酸主要组成成分。胁迫条件下,菌体脂肪酸发生不同程度变化,其中饱和脂肪酸即十四碳异脂肪酸(iso-C14:0)、十五碳异脂肪酸(iso-C15:0)和十八碳顺式单不饱和脂肪酸(cis-C18:1)从无到有,而iso-C16:1、anteiso-C17:0从有到无。说明二氧化硫胁迫不仅导致菌体脂肪酸含量发生变化,也调整了脂肪酸的组成成分。
表 2 不同二氧化硫胁迫下植物乳杆菌R23细胞膜磷脂脂肪酸相对含量Table 2. Relative contents of PLFA in L. plantarum R23 under SO2 stress磷脂脂肪酸
PLFA磷脂脂肪酸相对含量
Relative amount of PLFA/%胁迫前
Before stress胁迫后
After stress十二碳异脂肪酸 12:0 iso 1.36±0.08 2.45±0.11 十四烷酸 14:0 7.34±0.21 3.36±0.31 十四碳异脂肪酸 14:0 iso 0 1.92±0.18 十五碳异脂肪酸 15:0 iso 0 0.98±0.04 十六碳异脂肪酸 16:1 iso 3.99±0.18 0 十六碳顺式单不饱和脂肪酸 16:1 cis 3.19±0.55 2.87±0.65 十六烷酸 16:0 27.77±1.29 30.10±1.20 十七碳前异脂肪酸 17:0 anteiso 1.55±0.10 0 Sum In Feature 8 7.35±0.23 12.05±0.92 Sum In Feature 9 18.67±0.11 8.14±0.96 十八烷酸 18:0 8.65±0.59 10.92±0.17 十八碳顺式单不饱和脂肪酸 18:1 cis 0 2.46±0.20 十九碳反式脂肪酸 19:1 trans 2.41±0.16 2.29±0.40 Sum In Feature 11 4.28±0.03 7.80±0.08 环丙烷脂肪酸 19:0 cyclo 13.44±0.31 14.66±0.34 ![]() 图 4 二氧化硫对植物乳杆菌R23细胞膜磷脂脂肪酸相对含量的影响a、b、c、d分别表示二氧化硫胁迫导致的总饱和脂肪酸与总不饱和脂肪酸、总长链脂肪酸与总短链脂肪酸、总直链脂肪酸与总支链脂肪酸、总顺式脂肪酸与总反式脂肪酸的变化;“*”表示两者间的差异显著(P < 0.05)。Figure 4. Effect of SO2 on relative contents of PLFA in L. plantarum R23a, b, c, and d: changes caused by SO2 stress on saturated (Sa-), unsaturated (Us-), long-chain (L-), short-chain (S-), straight-chain (Sc-), branched-chain (Bc-), cis (Cis-), and trans fatty acids (Trans-) PLFAs. “*” indicates significant difference at P<0.05.
图 4 二氧化硫对植物乳杆菌R23细胞膜磷脂脂肪酸相对含量的影响a、b、c、d分别表示二氧化硫胁迫导致的总饱和脂肪酸与总不饱和脂肪酸、总长链脂肪酸与总短链脂肪酸、总直链脂肪酸与总支链脂肪酸、总顺式脂肪酸与总反式脂肪酸的变化;“*”表示两者间的差异显著(P < 0.05)。Figure 4. Effect of SO2 on relative contents of PLFA in L. plantarum R23a, b, c, and d: changes caused by SO2 stress on saturated (Sa-), unsaturated (Us-), long-chain (L-), short-chain (S-), straight-chain (Sc-), branched-chain (Bc-), cis (Cis-), and trans fatty acids (Trans-) PLFAs. “*” indicates significant difference at P<0.05.对不同类型脂肪酸的统计分析发现,二氧化硫胁迫后菌体总饱和脂肪酸含量上升,总不饱和脂肪酸含量下降(图4-a),两者比值提高1.75,即二氧化硫胁迫导致菌体膜脂肪酸饱和程度增加。类似的,二氧化硫胁迫导致了总直链脂肪酸提升(从49.36%升至52.0%),而总支链脂肪酸显著下降(从6.90%降至5.35%,P<0.05),总体直链/支链比值显著提升(从7.15升至9.72,P<0.05);值得注意的是,支链脂肪酸中的异脂肪酸虽未发生变化,但前异脂肪酸从1.55%降低为0,因此,总体异脂肪酸和前异脂肪酸的比值增大(图4-c)。另外,顺式/反式脂肪酸的比值也从1.33显著提升到2.33(P<0.05)(图4-d)。不同的是,菌体长链脂肪酸和短链脂肪酸在二氧化硫胁迫后的相对含量均有微量提升,但长链脂肪酸的增加幅度更大,因此总体上长链/短链的比值提升0.12(图4-b)。另外还可以发现,作为植物乳杆菌R23脂肪酸主要组成成分之一的环丙烷脂肪酸cyclo-C19:0,在胁迫处理后其相对含量也得以提升,提示其在该菌体耐受二氧化硫胁迫过程中发挥了一定的作用。从整体变化趋势分析,二氧化硫胁迫后植物乳杆菌R23膜脂肪酸成分向着饱和、长链、直链和环型调整。
3. 讨论与结论
3.1 抗氧化酶应激酶系对二氧化硫胁迫的响应
微生物体会启动自身应激机制以抵御各种逆境胁迫等不利条件,如产生特殊保护功能的蛋白[10]、改变细胞膜流动性[11-12]等,从而维持菌体正常的生长代谢和功能;本研究则发现植物乳杆菌R23通过提高抗氧化酶活力、调节细胞膜脂肪酸结构的方式来应对二氧化硫胁迫。已有研究发现,在生物体氧化应激反应中SOD、GPX和CAT是清除ROS的重要抗氧化酶类 [13],如将sod基因进行外源表达后,可明显提高乳杆菌的抗氧化能力[14];而发酵乳杆菌ME-3中的谷胱甘肽过化物酶和谷胱甘肽还原酶,可以从环境中运输及合成谷胱甘肽来参与抗氧化反应[15]。本研究植物乳杆菌R23抗氧化酶活力的显著提升表明该菌也启动了氧化应激反应,且其抗氧化系统整体呈现一个主动防御到被破坏的趋势。起始阶段,随着胁迫程度增大,SOD、GPX和CAT活性明显提高(P<0.05),以清除胞内过量的∙O2ˉ、H2O2、∙OH;但当二氧化硫质量浓度超过菌体耐受阈值时,各酶活力开始下降,MDA含量上升幅度加大。这揭示了过高二氧化硫胁迫会导致菌体抗氧化系统失调,使得胞内自由基产生和清除失去动态平衡,过量自由基与细胞膜中的脂肪酸等物质发生脂质过氧化反应,进而引起细胞膜流动性或通透性改变,促使胞内离子和其他电解质及可溶性物质大量外渗,破坏了细胞内酶及代谢作用原有的区域性,最终导致细胞皱缩甚至破裂。因此,抗氧化酶活性对菌体细胞应对氧化损伤起到至关重要的作用。
3.2 细胞膜对二氧化硫胁迫的响应
细胞膜是把菌体细胞与外界环境隔离的第一道屏障,也是逆境因子生理胁迫的首要目标之一,因此在维持胞内微环境的稳定性等方面发挥极其重要的作用[16]。一般认为,细胞膜的生化特性很大程度上取决于组成磷脂的脂肪酸结构[17]。本研究借助MIDI系统对二氧化硫胁迫前后植物乳杆菌R23膜脂肪酸测定结果发现,胁迫后菌体脂肪酸成分及比例均发生变化,整体向着饱和、长链、直链和环型增长,其中直链/支链、顺式/反式的比值显著提升(P<0.05)。由于总直链脂肪酸占比较高(50%以上),因此推断直链(尤其是C16:0)与支链的比例在植物乳杆菌R23防御二氧化硫胁迫中起主导作用。虽然本研究中菌体总饱和脂肪酸和长链脂肪酸含量增加不显著,但其占比也较大,所以认为它们同样发挥了积极作用。另有研究认为,不同类型脂肪酸的比例均会对细胞膜的流动性产生影响[18],其中支链脂肪酸(尤其是前异脂肪酸)中的甲基会破坏酰基链的紧凑性[19],降低膜流动性;如单增李斯特菌依靠增加直链脂肪酸同时降低前异脂肪酸C15:0、C17:0来适应酸性环境[20],而本研究中总支链脂肪酸增加以及前异脂肪酸anteiso-C17:0显著降低(P<0.05)应是植物乳杆菌R23应对二氧化硫胁迫的又一适应性调节方式。另外,长链饱和脂肪酸可以增加磷脂双分子层间酰基链的相互作用力,使其结构更为牢固且具有较高的相变温度和较低的渗透性[17];因此植物乳杆菌R23对长链/短链、饱和/不饱和脂肪酸的调整,可以降低细胞膜对HSO3−、SO32−等的透过性,从而防止ROS大量产生。另外,由于不饱和脂肪酸的氢键易被ROS氧化,本研究中其含量降低可以减弱膜脂对过氧化作用的敏感性。然而,目前关于不同类型脂肪酸在菌株抵御逆境胁迫中的作用并未取得一致观点。如:郑昀昀等[21]在新物种Anoxybacillus flavithermus ssp. Yunnanesis E13T对甲苯的耐受性,以及Huang 等[11]对植物乳杆菌ZDY2013 对酸的耐受性研究中,均证实饱和脂肪酸发挥了积极作用,与本研究结果相一致;而Taranto等[22]在罗伊氏乳杆菌耐胆盐、袁峥[23]在嗜酸乳杆菌耐酸的相关研究中发现,不饱和脂肪酸比例增加对菌体耐受性更有利;由此可见,不同特性的菌株或不同因子胁迫下其耐受性机制也不尽相同。
综上,本研究发现了植物乳杆菌R23抗氧化酶、膜磷脂脂肪酸等在其应答二氧化硫引发的氧化应激中的作用,可初步阐明该菌对二氧化硫胁迫的适应性生理机制,为深入理解该菌抵抗高二氧化硫环境而生存的分子机制提供理论依据,同时为筛选高抗逆性菌株以解决目前高酸性果酒降酸中存在的问题提供思路。
-
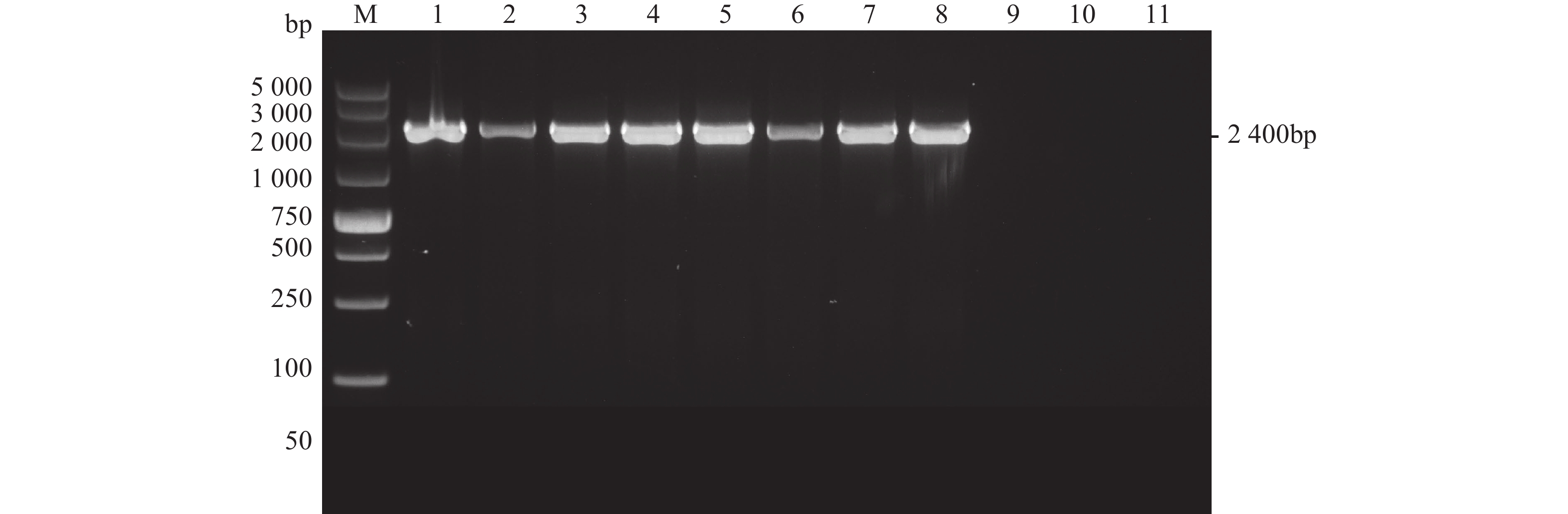
图 2 变形假单胞菌gyrB PCR扩增结果
M:Marker;1、变形假单胞菌H17050402;2、变形假单胞菌H17052503;3、变形假单胞菌H17041302;4、变形假单胞菌H16042701;5、变形假单胞菌ATCC700383;6、变形假单胞菌H15122403;7、变形假单胞菌H16040803;8、变形假单胞菌H16041203;9、溶藻弧菌H21112601;10、哈维氏弧菌H21112606:11、空白。
Figure 2. PCR on gyrB of P. plecoglossicida
M: Marker; 1: P. plecoglossicida H17050402; 2: P. plecoglossicida H17052503; 3: P. plecoglossicida H17041302; 4: P. plecoglossicida H16042701; 5: P. plecoglossicida ATCC700383; 6: P. plecoglossicida H15122403; 7: P. plecoglossicida H16040803; 8: P. plecoglossicida H16041203; 9: V. alginolyticus H21112601; 10: V. harveyi H21112606; 11: blank.
表 1 本研究用菌株及参考菌株
Table 1 Bacterial strains and reference used
菌株编号
Strains种属
Species来源地
Locations物种
Hosts分离部位
Tissues分离时间
DatesH15122403 变形假单胞菌
Pseudomonas plecoglossicida福建福鼎沙埕(沙埕港) 大黄鱼 脾 2015-12-24 H16040803 变形假单胞菌
Pseudomonas plecoglossicida福建霞浦东安(东吾洋) 大黄鱼 脾 2016-04-08 H16041203 变形假单胞菌
Pseudomonas plecoglossicida福建蕉城大湾(三都澳) 大黄鱼 脾 2016-04-12 H16042701 变形假单胞菌
Pseudomonas plecoglossicida福建蕉城黄湾(三都澳) 大黄鱼 肝 2016-04-27 H17041302 变形假单胞菌
Pseudomonas plecoglossicida福建蕉城泥土沃(三都澳) 大黄鱼 肾 2017-04-13 H17050402 变形假单胞菌
Pseudomonas plecoglossicida福建蕉城白秤潭(三都澳) 大黄鱼 肾 2017-05-04 H17052503 变形假单胞菌
Pseudomonas plecoglossicida福建蕉城白秤潭(三都澳) 大黄鱼 脾 2017-05-25 ATCC700383 变形假单胞菌
Pseudomonas plecoglossicida日本德岛县 香鱼 肾 1994-03 H13032003 荧光假单胞菌
Pseudomonas fluoresenes福建蕉城大湾(三都澳) 大黄鱼 脾 2013-03-20 H21112606 溶藻弧菌
Vibrio alginolyticus福建蕉城斗姆(官井洋) 大黄鱼 体表 2021-11-26 H21112601 哈维氏弧菌
Vibrio harveyi福建蕉城斗帽(官井洋) 大黄鱼 肝 2011-11-26 表 2 各菌株O抗原与抗O抗原血清的凝集反应结果
Table 2 Agglutination of O antigen and anti-O antigen sera
O抗原
O antigen抗O抗原血清凝集效价
Serum anti O antigen agglutination titerH15122403 H16040803 H16041203 H16042701 H17041302 H17050402 H17052503 H15122403 28 29 29 29 29 29 29 H16040803 28 29 28 28 28 28 28 H16041203 27 28 29 28 28 28 28 H16042701 28 29 29 28 27 29 28 H17041302 27 28 29 27 27 27 27 H17050402 27 28 29 26 27 29 28 H17052503 27 28 29 28 28 28 28 ATCC700383 27 28 28 29 28 28 29 H13032003 — — — — — — — H21112606 — — — — — — — H21112601 — — — — — — — 0.9%NaCl — — — — — — — 表 3 各菌株H抗原与抗H血清的凝集反应结果
Table 3 Agglutination of H antigen and anti-H antigen sera
H抗原
H antigen抗H抗原血清凝集效价
Serum anti H antigen agglutination titerH15122403 H16040803 H16041203 H16042701 H17041302 H17050402 H17052503 H15122403 29 29 29 29 29 28 28 H16040803 29 29 29 29 29 28 28 H16041203 29 29 29 29 29 29 29 H16042701 29 29 29 29 29 29 28 H17041302 29 29 29 29 29 29 29 H17050402 29 29 28 28 29 29 29 H17052503 29 29 28 28 29 28 29 ATCC700383 29 28 29 29 29 28 29 H13032003 — — — — — — — H21112606 — — — — — — — H21112601 — — — — — — — 0.9%NaCl — — — — — — — 表 4 其他临床分离菌株与7株变形假单胞菌抗血清的凝集结果
Table 4 Agglutination between individual strains and sera of 7 strains of P. plecoglossicida
抗原
Antigen抗O抗原血清/抗H抗原血清凝集效价
Serum anti O antigen/ Serum anti H antigen agglutination titer菌株
Strains时间
Dates来源
LocationsH15122403 H16040803 H16041203 H16042701 H17041302 H17050402 H17052503 H13032002 2013.03.20 宁德蕉城(三都澳) 28/29 28/29 28/28 29/29 27/27 26/26 27/28 H14060603 2014.06.06 宁德蕉城(三都澳) 29/28 29/28 29/27 29/28 29/29 29/28 29/28 H15012203 2015.01.22 宁德蕉城(三都澳) 29/29 29/28 29/29 29/29 28/28 29/28 29/29 H16031803 2016.03.18 宁德霞浦(东吾洋) 29/29 29/28 29/29 29/29 28/29 27/28 28/29 H16032302 2016.03.23 宁德福鼎(沙埕港) 29/28 28/29 27/29 28/29 29/29 28/29 28/28 H16032303 2016.03.23 宁德蕉城(三都澳) 29/28 29/28 29/27 28/29 29/28 28/29 29/29 H16042703 2016.04.27 宁德蕉城(三都澳) 29/28 29/29 29/29 29/29 29/28 29/28 28/29 H17030202 2017.03.02 宁德蕉城(三都澳) 29/29 29/29 29/28 28/29 29/29 28/29 28/28 H19040303 2019.04.03 宁德霞浦(东吾洋) 29/28 28/29 29/28 29/29 28/29 29/29 28/28 H20031102 2020.03.11 宁德蕉城(三都澳) 29/29 29/29 29/28 28/29 28/29 27/29 29/29 H22042903 2022.04.29 宁德蕉城(三都澳) 29/29 29/29 28/28 27/28 29/29 29/29 29/29 0.9%NaCl -/- -/- -/- -/- -/- -/- -/- -
[1] 农业农村部渔业渔政管理局, 全国水产技术推广总站, 中国水产学会. 中国渔业统计年鉴-2021[M]. 北京: 中国农业出版社, 2021: 22. [2] 谢书秋, 刘振勇. 闽东大黄鱼养殖现状分析与发展对策 [J]. 福建水产, 2006, 28(3):95−97. DOI: 10.3969/j.issn.1006-5601.2006.03.027 XIE S Q, LIU Z Y. Analysis of current situation and development countermeasures of Pseudosciaena crocea culture in East Fujian [J]. Journal of Fujian Fisheries, 2006, 28(3): 95−97.(in Chinese) DOI: 10.3969/j.issn.1006-5601.2006.03.027
[3] 张杰, 毛芝娟. 大黄鱼内脏白点病病原杀香鱼假单胞菌及其毒力因子研究进展 [J]. 浙江万里学院学报, 2015, 28(6):69−76,81. DOI: 10.3969/j.issn.1671-2250.2015.06.013 ZHANG J, MAO Z J. Research progress on Pseudomonas plecoglossicida and its virulence factors of large yellow croaker(Pseudosciaena crocea) [J]. Journal of Zhejiang Wanli University, 2015, 28(6): 69−76,81.(in Chinese) DOI: 10.3969/j.issn.1671-2250.2015.06.013
[4] 胡娇, 张飞, 徐晓津, 等. 大黄鱼(Pseudosciaena crocea)内脏白点病病原分离鉴定及致病性研究 [J]. 海洋与湖沼, 2014, 45(2):409−417. DOI: 10.11693/hyhz20140300078 HU J, ZHANG F, XU X J, et al. Isolation, identification and virulence of the pathogen of white-spots disease in internal organs of Pseudosciaena crocea [J]. Oceanologia et Limnologia Sinica, 2014, 45(2): 409−417.(in Chinese) DOI: 10.11693/hyhz20140300078
[5] 赖福才, 汪能平, 耿穗娜, 等. 应用铜绿假单胞菌4种分型方法的探讨 [J]. 中华医院感染学杂志, 2001, 11(2):81−83. DOI: 10.3321/j.issn:1005-4529.2001.02.001 LAI F C, WANG N P, GENG S N, et al. Investigation of four typing methods for Pseudomonas aeruginosa [J]. Chinese Journal of Nosoconmiology, 2001, 11(2): 81−83.(in Chinese) DOI: 10.3321/j.issn:1005-4529.2001.02.001
[6] 李献梅, 王小芬, 杨洪岩, 等. 促旋酶(gyrase)B亚单位基因gyrB在鉴别细菌近缘种中的应用 [J]. 微生物学报, 2008, 48(5):701−706. DOI: 10.3321/j.issn:0001-6209.2008.05.023 LI X M, WANG X F, YANG H Y, et al. Application of gyrB in the identification of closely related bacteria—a review [J]. Acta Microbiologica Sinica, 2008, 48(5): 701−706.(in Chinese) DOI: 10.3321/j.issn:0001-6209.2008.05.023
[7] 郝云婕, 韩素贞. gyrB基因在细菌系统发育分析中的应用 [J]. 生物技术通报, 2008(2):39−41. DOI: 10.13560/j.cnki.biotech.bull.1985.2008.02.007 HAO Y J, HAN S Z. Application of gyrB gene in bacterial phylogenetic analysis [J]. Biotechnology Bulletin, 2008(2): 39−41.(in Chinese) DOI: 10.13560/j.cnki.biotech.bull.1985.2008.02.007
[8] 高阳, 李璐璐, 骆延波, 等. 铜绿假单胞菌的血清分型、鞭毛分型和脉冲场凝胶电泳分型 [J]. 中国兽医学报, 2018, 38(2):341−348. DOI: 10.16303/j.cnki.1005-4545.2018.02.19 GAO Y, LI L L, LUO Y B, et al. Serotyping, flagella typing and pulse field gel electrophoresis of Pseudomonas aeruginosa isolates [J]. Chinese Journal of Veterinary Science, 2018, 38(2): 341−348.(in Chinese) DOI: 10.16303/j.cnki.1005-4545.2018.02.19
[9] 张嵘, 蔡加昌, 张书梅, 等. gyrB基因和16S rRNA基因序列分析在沙门菌属细菌鉴别中的临床应用评价 [J]. 中华微生物学和免疫学杂志, 2007, 27(4):368−369. DOI: 10.3760/j:issn:0254-5101.2007.04.020 ZHANG R, CAI J C, ZHANG S M, et al. Evaluation of clinical application of gyrB gene and 16S rRNA gene sequence analysis in the identification of Salmonella [J]. Chinese Journal of Microbiology and Immunology, 2007, 27(4): 368−369.(in Chinese) DOI: 10.3760/j:issn:0254-5101.2007.04.020
[10] 许斌福, 龚晖, 李素一, 等. 鳗源创伤弧菌的鉴定与血清型分析 [J]. 渔业研究, 2016, 38(5):351−356. XU B F, GONG H, LI S Y, et al. Identification and serotypes analysis of Vibrio vulnificus from Anguilla anguilla [J]. Journal of Fisheries Research, 2016, 38(5): 351−356.(in Chinese)
[11] 赵建梅, 李月华, 宋传周, 等. PCR鉴定沙门氏菌血清分型方法的建立与应用 [J]. 中国动物检疫, 2018, 35(1):73−77. DOI: 10.3969/j.issn.1005-944X.2018.01.021 ZHAO J M, LI Y H, SONG C Z, et al. Establishment and application of PCR assay for serotype identification of Salmonella [J]. China Animal Health Inspection, 2018, 35(1): 73−77.(in Chinese) DOI: 10.3969/j.issn.1005-944X.2018.01.021
[12] 吴雨辰, 胡燕燕, 张嵘. 细菌分型方法的研究进展 [J]. 中国感染与化疗杂志, 2021, 21(5):617−623. DOI: 10.16718/j.1009-7708.2021.05.021 WU Y C, HU Y Y, ZHANG R. Advances in bacterial typing methods [J]. Chinese Journal of Infection and Chemotherapy, 2021, 21(5): 617−623.(in Chinese) DOI: 10.16718/j.1009-7708.2021.05.021
[13] NISHIMORI E, KITA-TSUKAMOTO K, WAKABAYASHI H. Pseudomonas plecoglossicida sp. nov. , the causative agent of bacterial haemorrhagic ascites of ayu, Plecoglossus altivelis[J]. International Journal of Systematic and Evolutionary Microbiology, 2000, 50(Pt 1): 83-89.
[14] IZUMI S, YAMAMOTO M, SUZUKI K, et al. Identification and detection of Pseudomonas plecoglossicida isolates with PCR primers targeting the gyrB region [J]. Journal of Fish Diseases, 2007, 30(7): 391−397. DOI: 10.1111/j.1365-2761.2007.00820.x
[15] 王晓婷. 7株变形杆菌的O抗原基因簇的破译和特异基因的鉴定[D]. 天津: 南开大学, 2011. WANG X T. Specific gene identification and O antigen gene clusters analysis of 7 Proteus[D]. Tianjin: Nankai University, 2011.
[16] 李可, 方莹, 张晓峰, 等. 沙门氏菌的血清分型及分子鉴定研究进展 [J]. 食品安全质量检测学报, 2016, 7(10):3947−3951. DOI: 10.19812/j.cnki.jfsq11-5956/ts.2016.10.017 LI K, FANG Y, ZHANG X F, et al. Research progress of Salmonella serotyping and molecular identification methods [J]. Journal of Food Safety & Quality, 2016, 7(10): 3947−3951.(in Chinese) DOI: 10.19812/j.cnki.jfsq11-5956/ts.2016.10.017
[17] 郭维植, 程法稷, 林成水, 等. 能与大肠杆菌O157(H7)抗血清交叉凝集的5种细菌的分离和鉴定 [J]. 海峡预防医学杂志, 2000, 6(4):4−6. DOI: 10.3969/j.issn.1007-2705.2000.04.002 GUO W Z, CHENG F J, LIN C S, et al. Isolation and identification of 5 species bacteria cross reacting with E. coli O157 (H7) anti serums [J]. Strail Journal of Preventive Medicine, 2000, 6(4): 4−6.(in Chinese) DOI: 10.3969/j.issn.1007-2705.2000.04.002
[18] 邵西群, 胡博, 章秀婷, 等. 水貂出血性肺炎病原铜绿假单胞菌的分子特征 [J]. 中国兽医科学, 2014, 44(4):340−345. DOI: 10.16656/j.issn.1673-4696.2014.04.001 SHAO X Q, HU B, ZHANG X T, et al. Molecular characterization of Pseudomonas aeruginosa strains isolated from minks with haemorrhagic pneumonia [J]. Chinese Veterinary Science, 2014, 44(4): 340−345.(in Chinese) DOI: 10.16656/j.issn.1673-4696.2014.04.001
[19] 宋程, 陈小玲. PCR技术用于病原微生物的血清型鉴定研究进展 [J]. 中国兽药杂志, 2001, 35(1):59−62. DOI: 10.3969/j.issn.1002-1280.2001.01.015 SONG C, CHEN X L. Research progress of PCR technology in serotype identification of pathogenic microorganisms [J]. Chinese Journal of Veterinary Drug, 2001, 35(1): 59−62.(in Chinese) DOI: 10.3969/j.issn.1002-1280.2001.01.015




 下载:
下载: