Cloning and Expression of DcbHLH14 from Dendrobium catenatum Lindl.
-
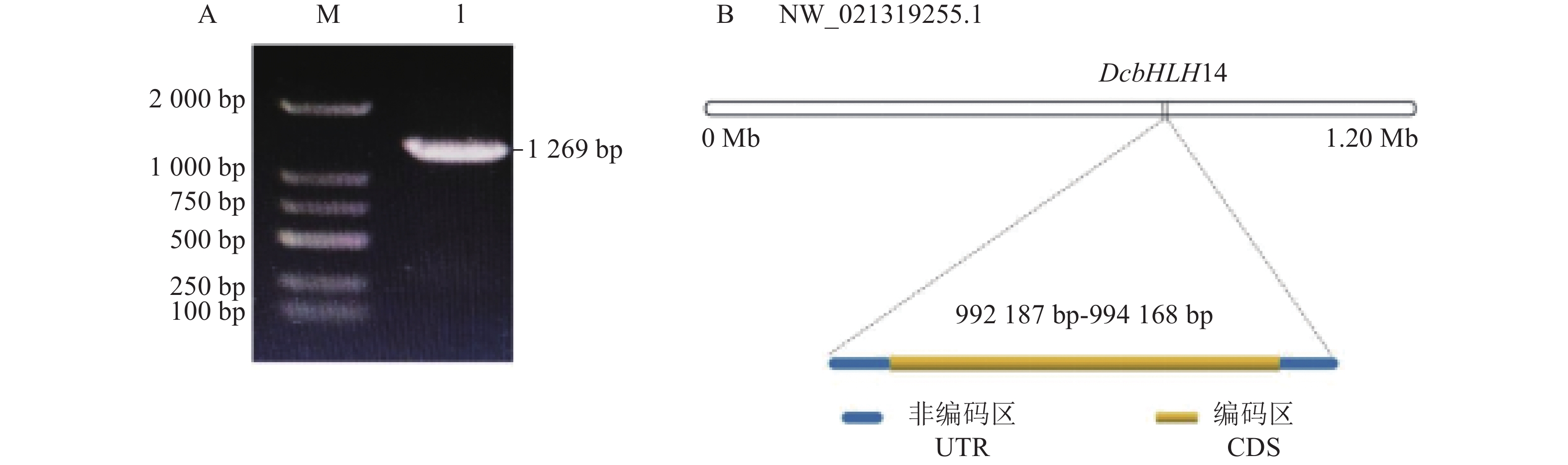
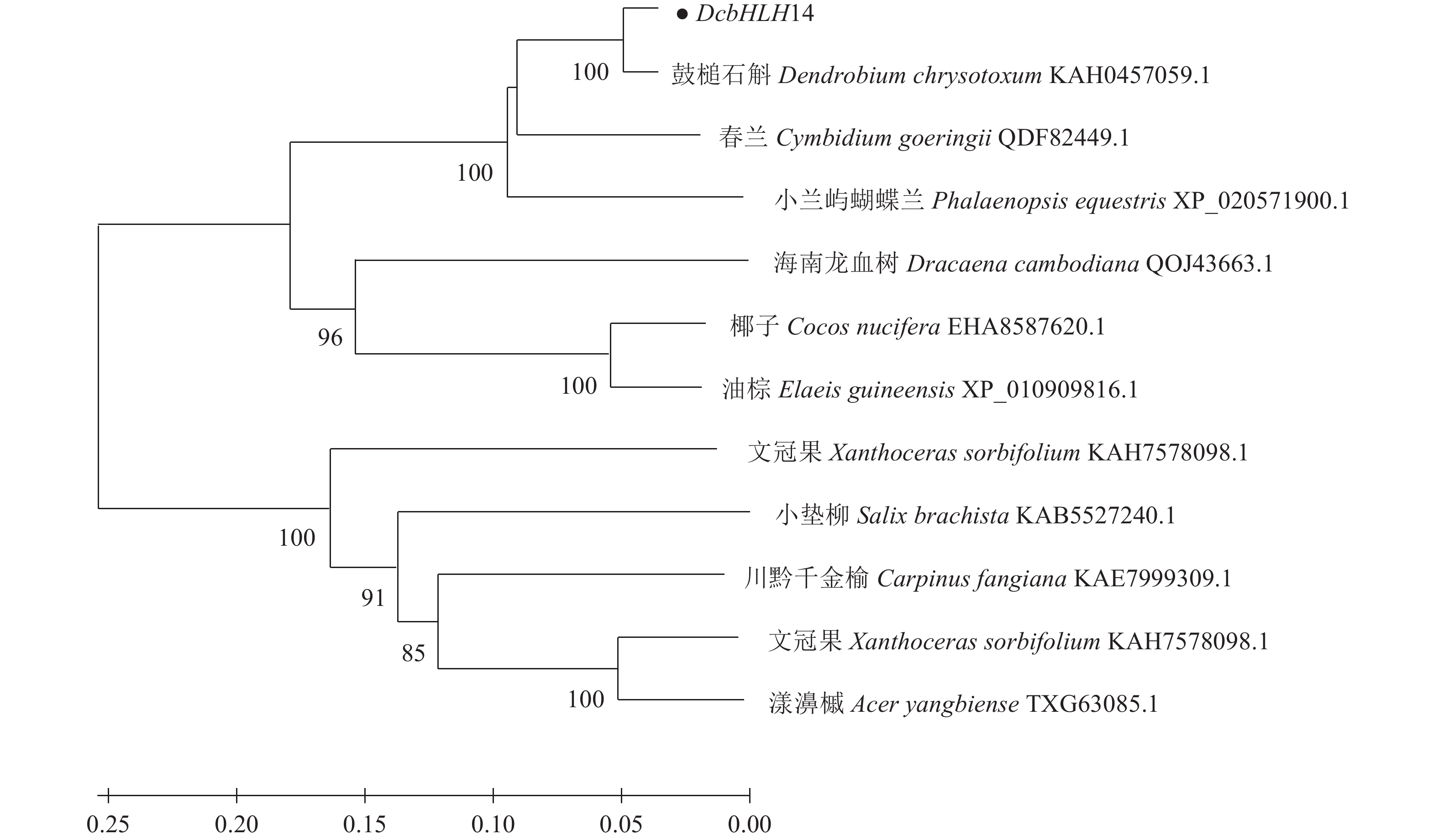
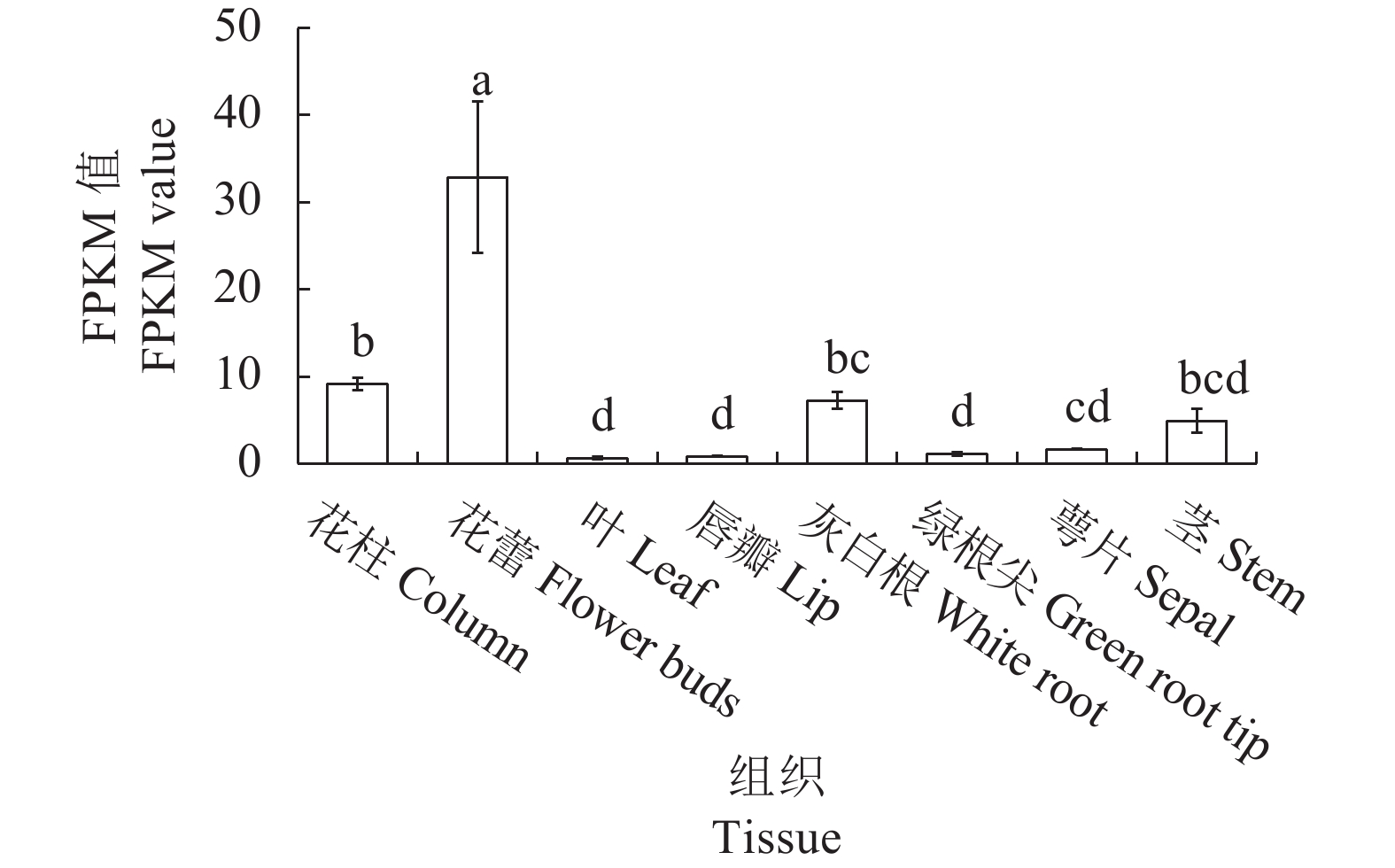
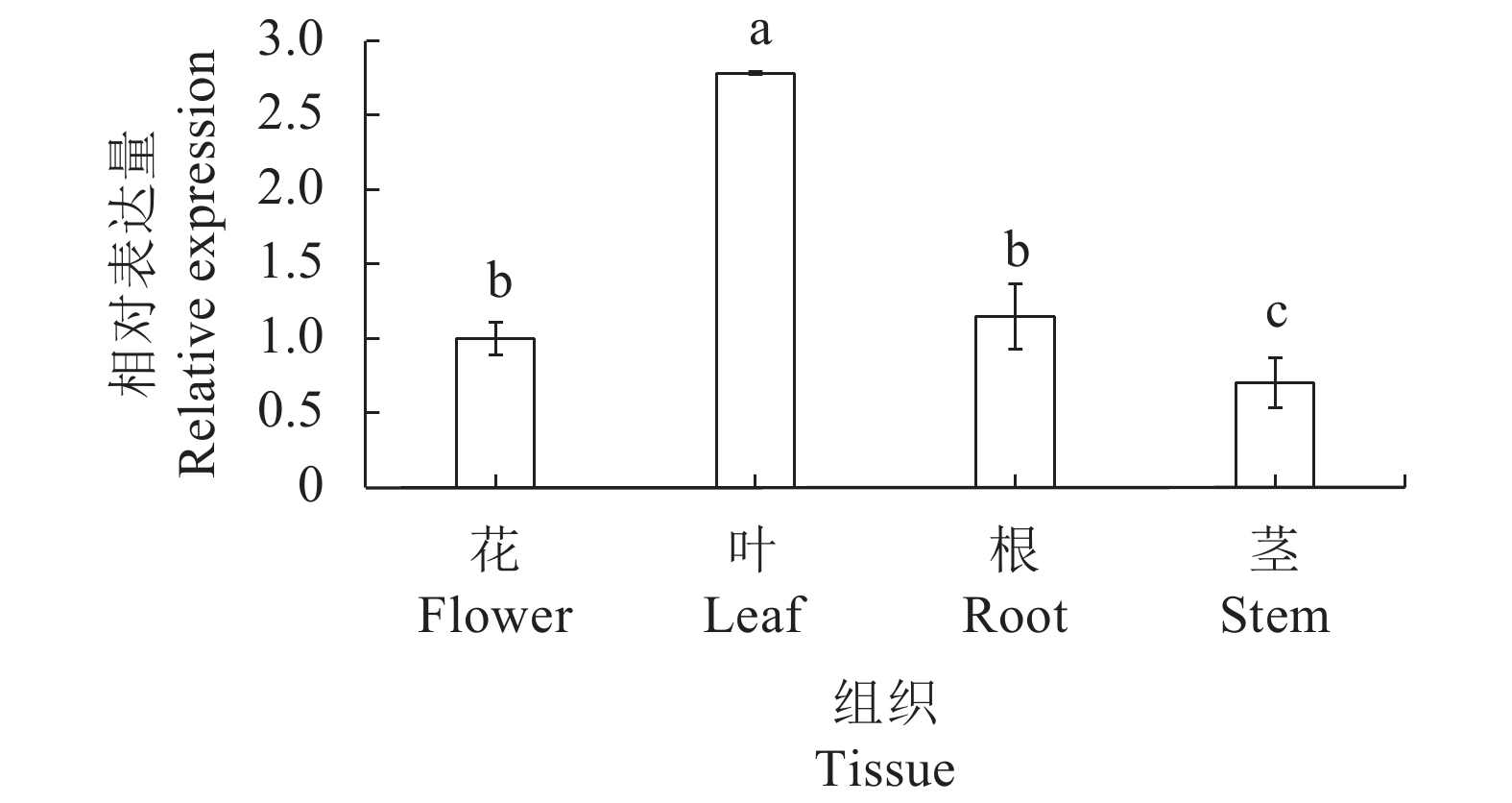
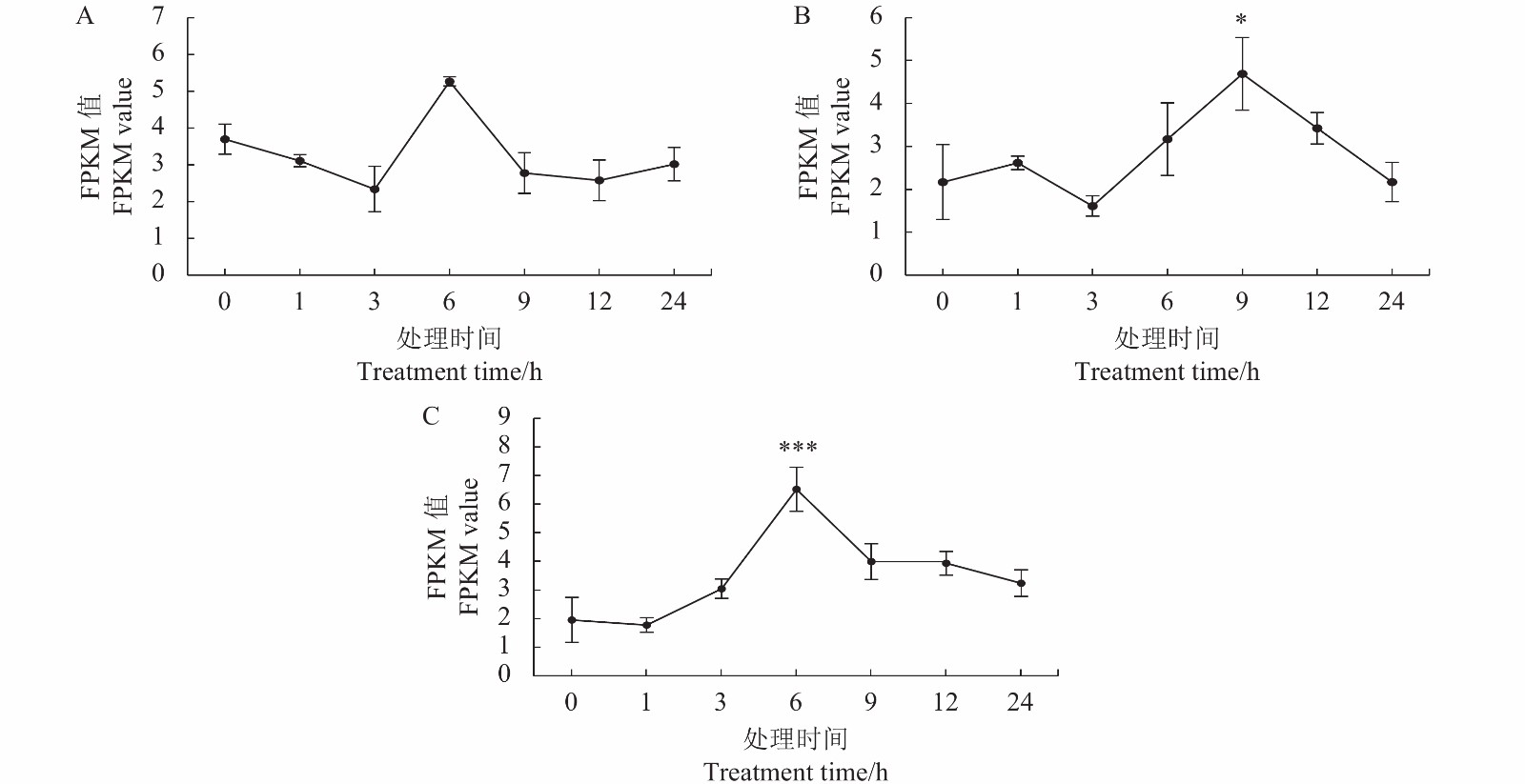
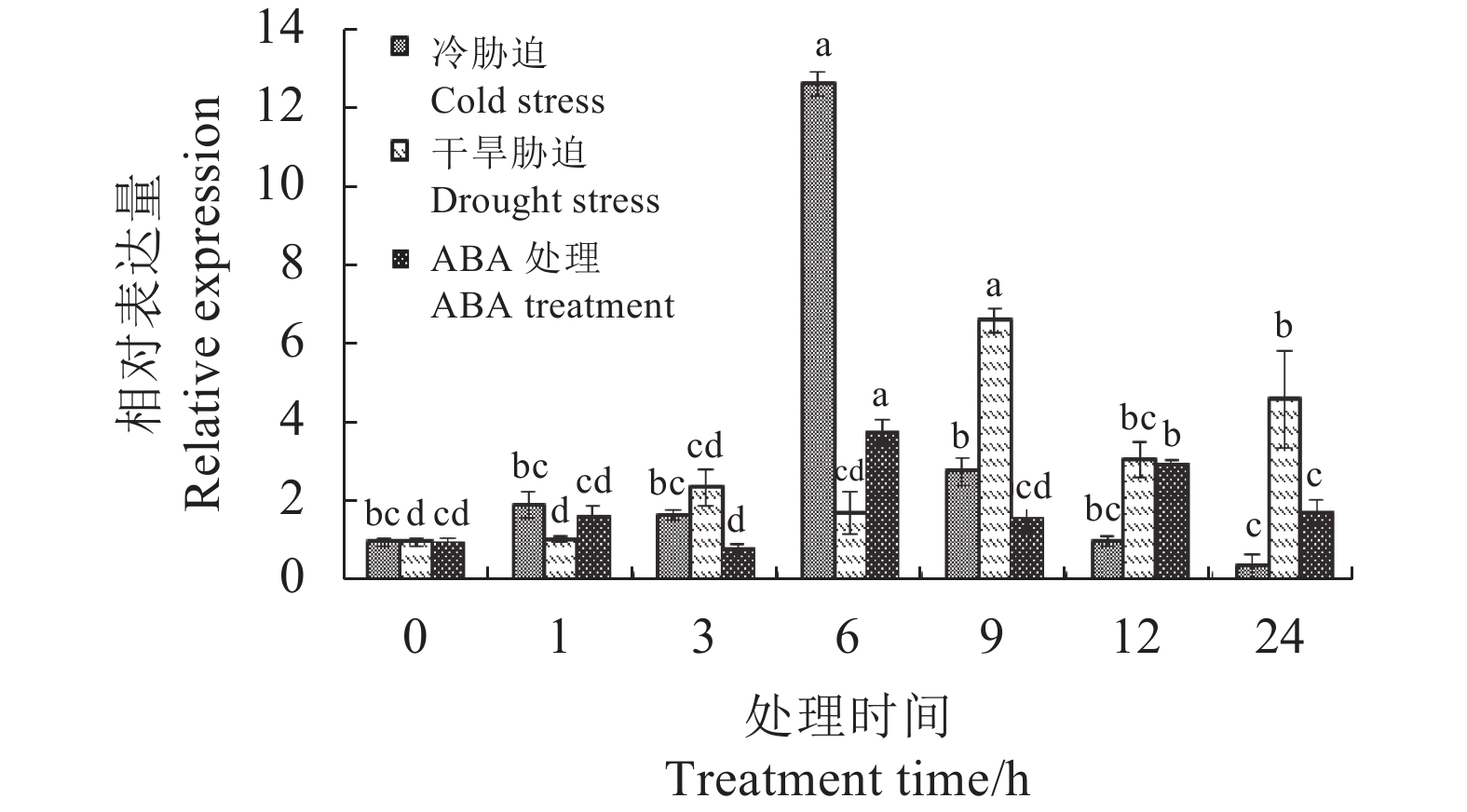
摘要:目的 克隆铁皮石斛转录因子DcbHLH14基因并分析其在非生物胁迫响应中的表达情况,为研究DcbHLH14基因的功能提供理论参考。方法 通过同源克隆从铁皮石斛叶组织中得到DcbHLH14基因,对其编码的蛋白序列特征和组织表达特性进行分析,并采用qRT-PCR对DcbHLH14基因在低温、干旱和ABA处理过程中的表达量进行分析。结果 DcbHLH14基因开放阅读框(Open reading frame, ORF)为1269 bp,与参考序列存在7个碱基差异,仅含有1个外显子且无内含子,编码422个氨基酸。DcbHLH14蛋白的分子式为C2011H3192N586O613S13,理论分子量为45.8 kD,理论等电点(pI)为5.98,含有bHLH家族保守结构域bHLH-MYC-N和HLH,属于bHLH家族,与鼓槌石斛(Dendrobium chrysotoxum)和春兰(Cymbidium goeringii)bHLH蛋白的氨基酸序列同源性较高,分别为97.16%和86.90%。转录组分析结果显示,DcbHLH14基因在云南产地野生铁皮石斛花蕾中的表达量最高,在叶中的表达量最低。进一步qRT-PCR分析结果表明,该基因在广东丹霞铁皮石斛叶中的表达量最高,而在茎中的表达量最低。DcbHLH14基因启动子富含多种与水分胁迫、低温、脱水以及ABA响应等相关的顺式作用元件。DcbHLH14基因明显受到低温、干旱和ABA诱导,低温和ABA处理6 h后DcbHLH14表达量被显著提高并达到峰值,分别是处理前的12.6倍和3.7倍;干旱处理9 h后,DcbHLH14表达量最高,是处理前的6.5倍,达到极显著差异水平。结论 DcbHLH14基因可能在转录水平上通过依赖于ABA信号通路途径响应低温和干旱胁迫,从而调控下游功能基因表达,提高铁皮石斛抗逆性。
-
关键词:
- 铁皮石斛 /
- DcbHLH14基因 /
- 非生物胁迫 /
- 表达分析
Abstract:Objective Functions of DcbHLH14, a basic helix-loop-helix transcription factor of Dendrobium catenatum Lindl., in response to abiotic stresses were studied.Method DcbHLH14 was cloned from D. catenatum leaves using homologous cloning method for a bioinformatic analysis on the gene and expression in tissues. Gene expressions under low temperature, drought, and abscisic acid (ABA) stresses were determined.Result The ORF of DcbHLH14 was 1 269 bp and encoded 422 amino acids. Itcontained one exon, no intron, and 7 bases that were different from the reference sequences. The theoretical molecular weight was 45.8 kD, isoelectric point pH 5.98, and molecular formula C2011H3192N586O613S13. Its conserved domains contained bHLH-MYC-N and HLH proteins that had high similarities with the bHLH proteins in D. chrysotoxum at 97.16% and in Cymbidium goeringii at 86.90%. The transcriptome analysis revealed high expressions in the flower buds and columns but low in the leaves of the wild D. catenatum from Yunnan, whereas the qRT-PCR analysis showed high expressions in the leaves and low in the stems of the sample from Danxia, Guangdong. The promoters of DcbHLH14 contained numerous cis-acting elements associated with the responses to water-depletion, low temperature, dehydration, and ABA stresses, which significantly affected expression of the gene. For instance, DcbHLH14 was upregulated to peak in 6 h after a low temperature or ABA treatment reaching 12.6 or 3.7 times, respectively, as well as by a 9 h drought stress to become as high as 6.5 times of control. Conclusion It was postulated that DcbHLH14 responded to low temperature or drought stress through the ABA signaling pathway at transcription level. Hence, the tolerance of D. catenatum to the abiotic stresses could be manipulated by regulating the expression of the downstream functional gene.-
Keywords:
- Dendrobium catenatum /
- DcbHLH14 /
- abiotic stress /
- expression analysis
-
0. 引 言
【研究意义】卷荚相思(Acacia cincinnata)属豆科金(Leguminosae)合欢(Vachellia farnesiana)属植物,在我国南方地区广泛种植。卷荚相思是沿海沙地重要的树种资源之一,具有适生性强、产量高、生长快、性状优良等特性,是优质的家具用材和造纸树种[1]。但在林业生产中,经常会出现种子空壳、质量不高、籽粒不饱满等现象,最终导致卷荚相思种子萌发率不高,因此选择合适的方法提高卷荚相思种子的萌发率非常必要。物理农林业是物理技术和农林业生产有机结合的一种生产方式,将电、磁、光、热等物理学科知识与农林业生产相关领域的前沿技术结合起来,使用特殊技术方法处理植物,在无污染的条件下,达到增产、优质、抗病和高效的目的,并且在工艺上和成本上都易于实现,在农林业中的应用和研究是十分广泛的。目前,电场种子处理技术应用于种植业和养殖业,在农林业生产和环境保护中已发挥巨大的作用[2]。为此探讨电场处理对卷荚相思种子萌发的影响效应,为研究种子萌发提供理论基础。【前人研究进展】相关研究表明,电场与时间两者交互处理不仅可以刺激植物种子萌发、生长、愈伤组织形成以及细胞融合,而且可以提高细胞膜内的电位差,膜内外电位差的改变,影响物质交换的速度[3];电场处理技术在食品加工和贮存[4]、畜牧业[5]、水产养殖业[6]和病虫害防治[7]等方面都获得了重要的成果,取得了显著的经济效益[8]。种子顺利萌发、成苗是植物完成生活史的前提,除受种子自身大小、质量、环境和储存方式等因素影响外,还受制于前期对种子的处理等重要因素[9]。利用适宜的电场处理植物种子可以明显提高种子活力,并且提高种子的发芽率、发芽势和发芽指数,使其出苗整齐和迅速,促进了作物对生长发育所需要养分的吸收,加速植物的生长发育,促进其光合作用,提高植物的产量及质量[10−15]。【本研究切入点】目前电场处理对卷荚相思种子萌发影响和生长的影响研究鲜见报道,电场强度和处理时间对植物种子萌发的影响阈值尚未确定。【拟解决的关键问题】开展不同电场强度和处理时间交互下对卷荚相思种子萌发的影响试验,研究萌发阈值及其对生长指标影响等问题,为卷荚相思种子萌发及可持续经营提供理论参考。
1. 材料与方法
1.1 试验区概况
试验地位于福建农林大学(119°13′51.73″E、26°04′55.5″N),属于亚热带季风性气候,全年冬暖夏凉,年平均日照1755.4 h,无霜期达360 d以上。年均温为19.9 ℃,极端最高温42 ℃,最低温度为0 ℃,年空气相对湿度79 %。
1.2 供试材料
试验所用卷荚相思种子于2022年3月采集于福建省漳浦中西国有林场,选取大小一致、颗粒饱满的种子用于萌发试验。
1.3 试验设计
本试验电场由波尔高压电源73030P型电场发生器产生,使用2块平行金属板进行连接且金属板之间保持0.5 cm距离并形成一个连续可调的电场,种子均匀分布在底层金属板上。利用二次通用旋转组合设计[16]进行预试验,设置电场强度分别为0.2、0.4、0.6、0.8、1.0 kV·cm−1和处理时间分别为15、30、45、60 min的电场处理,未经处理为对照(CK),以电压强度和时间为控制因素,共计20个处理,1个对照,每个处理3个重复,每个重复50粒种子。
1.4 试验方法
电场处理种子后第2天进行置床,采用纸上培养法[17]进行种子萌发试验,将处理后的种子用15% 福尔马林消毒30 min并用蒸馏水冲洗数遍。将各培养皿内滤纸润湿后每个培养皿50粒种子,并重复3次。将培养皿放入恒温培养箱中进行发芽试验,设定萌发条件为温度25 ℃,湿度70%,12 h光照和12 h黑暗交替。在每个培养皿中加入2 mL蒸馏水,每天17:00记录种子萌发情况。当种子胚根突破种皮2 mm时,视为种子萌发。
1.5 萌发指标计算
卷荚相思种子在第3天开始萌发并于12 d后结束发芽,发芽结束后分别计算发芽率、发芽势和发芽指数。公式如下[18]:
发芽率(GE)/% = (发芽种子数/供试种子数) ×100;
发芽势(GR)/% = (初期发芽种子数/供试种子数) ×100;
发芽指数 (GI) = ∑(第t天种子发芽数/相应发芽天数)。
1.6 数据分析
采用SPSS 25.0对种子萌发指标进行统计分析,运用Duncan’s法进行多重比较(P<0.05)不同电场和时间交互处理种子萌发特性;种子萌发指标采用平均值±标准误表示。为了验证数据是否满足条件,对其进行KS检验,如若原始数据不满足正态分布,对原始数据进行10作底数的lg(x+1)对数转换,保证数据呈正态分布。采用Origin 2022进行非线性回归分析和聚类分析。
2. 结果与分析
2.1 电压和时间交互处理对种子萌发特性的影响
2.1.1 不同电场强度和时间交互处理对种子发芽率的影响
在电场强度和时间交互处理下,卷荚相思的种子发芽率在一定程度上存在差异(表1)。从电场强度看,在15~60 min时发芽率随着电场的增高与CK相比呈现先降后升再降的趋势,且在电场强度0.6 kV·cm−1,处理时长为45、60 min时和电场强度0.8 kV·cm−1,处理时长为30 min时卷荚相思种子的发芽率均达较高水平,分别为75.00%、74.67%和74.33%,比CK分别提高13.64%、13.14%和12.62%,且存在显著差异。从处理时间来看,在电场强度为0.6 、0.8 kV·cm−1时,随着处理时间的延长,卷荚相思种子发芽率逐渐提高,呈现先升后降的趋势,且均高于CK,0.6 kV·cm−1时与CK相比分别提高2.5%、8.1%、13.63%和13.14%;0.8 kV·cm−1时与CK相比分别提高1.0%、12.62%、8.1%和6.1%。说明在电场强度0.8 kV·cm−1、处理时间45 min的双因素交互处理下,可以有效提高种子的发芽率。
表 1 不同电场强度与时间交互处理对卷荚相思种子发芽率的影响Table 1. Effect of applied electric field and treatment time on germination rate of A. cincinnata seeds(单位:%) 电场强度
Electric field strength/(kV·cm−1)处理时间 Handling time 0 min 15 min 30 min 45 min 60 min 0.0 66.00±4.00 aA 66.00±4.00 aA 66.00±4.00 aB 66.00±4.00 aC 66.00±4.00 aAB 0.2 66.00±4.00 aA 48.67±2.52 cC 55.00±2.00 bcD 66.00±3.00 aC 60.67±5.77 abB 0.4 66.00±4.00 aA 56.00±2.00 bB 67.33±1.15 aB 70.33±1.53 aAB 70.00±3.46 aA 0.6 66.00±4.00 bA 67.67±2.52 abA 71.33±3.06 abAB 75.00±3.00 aA 74.67±7.57 aA 0.8 66.00±4.00 cA 66.67±2.08 bcA 74.33±3.51 aA 71.33±2.52 abAB 70.00±2.00 abcA 1.0 66.00±4.00 aA 65.33±2.52 aA 60.33±3.51 abC 59.33±2.52 bD 50.00±2.00 cC 同行中不同小写字母代表不同处理时间存在显著差异(P<0.05),同列中不同大写字母代表不同电场强度存在显著差异(P<0.05)。下同。
Data with different lowercase letters on same row represent significant differences under different treatment times (P<0.05); and those with different uppercase letters on same column represent significant differences under different electric field (P<0.05). Same for below.2.1.2 不同电场强度和时间交互处理对种子发芽势的影响
不同电场强度和时间交互处理对卷荚相思种子发芽势有不同程度的影响且存在一定差异(表2)。不同强度的电场可产生不同的生物学效应,发芽势有时升高,有时降低,但整体呈现出促进作用。0.2 kV·cm−1处理下随着时间的延长呈现先下降后上升再下降的趋势,且在15 min时与CK有显著差异(P<0.05);0.4 kV·cm−1处理下随着时间的延长呈现先降后升的趋势,在60 min时发芽势达最高,但是与CK无显著差异(P>0.05);0.6~1.0 kV·cm−1 处理下随着时间的延长呈现先升后降的趋势,且电场强度在0.8 kV·cm−1、处理时间45 min时发芽势达到最高并且与CK相比有显著差异(P<0.05),表2可以看出电场强度在0.6 、0.8 kV·cm−1,处理时间在30~45 min时,对卷荚相思种子的发芽势有一定的促进作用,并且与CK相比显著提高(P<0.05)。说明电场处理对卷荚相思种子具有一定的改善作用,在电场强度0.8 kV·cm−1、处理时间45 min可以促进发芽势的提高。
表 2 不同电场强度与时间交互处理对卷荚相思种子发芽势的影响Table 2. Effect of applied electric field and treatment time on germination potential of A. cincinnata seeds(单位:%) 电场强度
Electric field strength/(kV·cm−1)处理时间 Handling time 0 min 15 min 30 min 45 min 60 min 0.0 39.67±2.08 aA 39.67±2.08 aCD 39.67±2.08 aB 39.67±2.08 aB 39.67±2.08 aB 0.2 39.67±2.08 abA 30.51±1.47 cE 36.24±3.31 abB 40.38±3.19 aB 35.33±1.15 bBC 0.4 39.67±2.08 abA 37.99±1.98 cD 38.38±4.65 cB 42.43±5.00 abB 46.67±4.16 aA 0.6 39.67±2.08 bA 48.02±1.60 aA 49.99±4.52 aA 50.58±1.87 aA 49.00±3.00 aA 0.8 39.67±2.08 cA 47.33±3.88 bAB 50.37±1.46 abA 52.73±1.79 aA 47.33±3.05 bA 1.0 39.67±2.08 abA 42.98±3.61 aBC 41.26±3.85 aB 41.23±5.27 aB 33.19±2.71 bC 2.1.3 不同电场强度和时间交互处理对种子发芽指数的影响
不同电场强度和时间交互处理对卷荚相思种子发芽指数有不同程度的影响并且存在一定的差异(表3)。从不同的电场强度看,0.2 kV·cm−1处理下的发芽指数在45 min时处于最高值,与CK相比提高了13.91%,但并无显著差异(P>0.05);0.4 kV·cm−1处理下的发芽指数呈现先降后升的趋势;0.6~1.0 kV·cm−1处理下的发芽指数均呈现先升后降的趋势,且在电场强度0.8 kV·cm−1、处理时间45 min时处于最高值,与CK相比提高了24.63%,具有显著差异(P<0.05);0.6 kV·cm−1处理下在处理时间15~60 min时与CK均有显著差异(P<0.05),1.0 kV·cm−1处理下在45 、60 min与CK相比降低了11.95%和12.68%,具有显著差异(P<0.05)。在相同处理时间下,随着电场强度的增强对发芽指数均产生不同程度的影响,可以看出0.4~0.6 kV·cm−1电场强度在30~60 min处理时间下对卷荚相思种子发芽指数刺激的作用优于0.2 、1.0 kV·cm−1的电场强度。从表中可以看出在电场和时间两者交互处理下能够促进卷荚相思种子的萌发,在电场强度0.8 kV·cm−1、处理时间45 min可以促进发芽指数的提高,具有优化促进作用。
表 3 不同电场强度与时间交互处理对卷荚相思种子发芽指数的影响Table 3. Effect of applied electric field and treatment time on germination index of A. cincinnata seeds电场强度
Electric field strength/(kV·cm−1)处理时间 Handling time 0 min 15 min 30 min 45 min 60 min 0.0 8.12±0.40 aA 8.12±0.40 aB 8.12±0.40 aBC 8.12±0.40 aCD 8.12±0.40 aBCD 0.2 8.12±0.40 abA 6.21±0.41 cD 7.38±0.63 bC 9.25±1.01 aABC 7.79±0.51 bCD 0.4 8.12±0.40 abA 7.12±0.47 bC 8.74±0.77 aAB 8.63±0.79 aBC 9.15±0.36 aAB 0.6 8.12±0.40 bA 9.24±0.66 abA 9.83±0.71 aA 9.57±0.55 abAB 9.35±1.18 abA 0.8 8.12±0.40 cA 8.91±0.22 bcAB 9.67±0.55 abA 10.12±0.52 aA 8.85±0.47 bcABC 1.0 8.12±0.40 aA 8.16±0.37 aB 7.64±0.70 abBC 7.15±0.23 bD 7.09±0.33 bD 2.1.4 电场和时间双因素交互处理对种子形态指标的影响
电场和时间交互处理对卷荚相思种子形态指标的双因素方差分析结果(表4)表明,电场、时间及两者交互处理对卷荚相思种子的发芽率、发芽势和发芽指数均具有显著影响(P<0.05)。
表 4 电场和时间交互处理对卷荚相思种子形态指标影响的双因素方差分析Table 4. Two-way ANOVA on effects of applied electric field and treatment time on morphological indexes of A. cincinnata seeds参数
Parameter电场强度
Electric field strength处理时间
Handling time电场强度×处理时间
electric field strength×
Handling timeF值
F valueP值
P valueF值
F valueP值
P valueF值
F valueP值
P value发芽率 GE 51.004 0.000 13.908 0.000 8.292 0.000 发芽势 GR 42.144 0.000 4.332 0.010 3.139 0.003 发芽指数 GI 27.919 0.000 7.365 0.000 4.294 0.000 2.2 卷荚相思种子萌发特性模型及分类分析
2.2.1 卷荚相思种子发芽率非线性回归方程的建立
对不同电场强度与处理时间进行非线性回归分析(表5)。发芽率(Y)作为因变量,自变量为电场(X1)和时间(X2),将这2个变量引入方程。通过非线性回归分析,不同电场强度与时间交互处理下的最优回归方程为:Y=7.73+135.23X1+1.24X2−86.16X12−0.00911X22−0.75X1X2,非线性回归方程达到显著水平(P<0.05),对回归方程进行显著性F检验,其F值为45.01806,大于F0.05(5,50)=2.40,表明非线性回归方程与实际情况拟合度高,能反映2项因素与发芽率的综合关系,说明上述模型具有统计学意义。
表 5 不同电场强度与时间交互处理对发芽率非线性回归模型Table 5. Nonlinear regression model between seed germination rate and applied electric field/time方程拟合度
R2调整后R2
Adjusted R2平方和 Square sum df 均方 Mean square F值 F value 回归
regression残差
residual error回归
regression残差
residual error回归
regression残差
residual error0.81 0.79 3251.04548 779.93786 5 54 650.2091 14.44329 45.01806 2.2.2 不同电场强度和时间交互处理下种子萌发特性热图及其聚类分析
为了直观分析5种电场强度在经过不同处理时间的交互作用,卷荚相思萌发期3个指标在相同环境下的变化情况,利用热图对卷荚相思种子萌发期性状的变化量进行可视化处理,通过纵向聚类反映3个指标在同一环境下的相互关系。5种电场强度处理下萌发期3个指标相对值的变化热图及聚类分析结果见图1。
![]() 图 1 不同电场强度与时间交互处理下的聚类分析①热图中红色表示电场对指标有促进作用,且颜色越深,作用越强;② t1-0.2、t2-0.2、t3-0.2、t4-0.2代表0.2 kV·cm−1,t1~t4分别代表处理时间为15、30、45和60 min,以此类推;③GE:发芽率,GR:发芽势,GI:发芽指数。Figure 1. Clustering of applied electric field and treatment time.①Red indicates how electric field promotes indicators--the darker the color, the stronger the effect; ② t1−0.2, t2−0.2, t3−0.2, and t4−0.2 represent treatments under 0.2kV·cm−1; t1-t4 represent treatments of 15, 30, 45, and 60m, respectively. Same for the others; ③GE stands for germination rate; GR stands for germination potential; GI stands for germination index.
图 1 不同电场强度与时间交互处理下的聚类分析①热图中红色表示电场对指标有促进作用,且颜色越深,作用越强;② t1-0.2、t2-0.2、t3-0.2、t4-0.2代表0.2 kV·cm−1,t1~t4分别代表处理时间为15、30、45和60 min,以此类推;③GE:发芽率,GR:发芽势,GI:发芽指数。Figure 1. Clustering of applied electric field and treatment time.①Red indicates how electric field promotes indicators--the darker the color, the stronger the effect; ② t1−0.2, t2−0.2, t3−0.2, and t4−0.2 represent treatments under 0.2kV·cm−1; t1-t4 represent treatments of 15, 30, 45, and 60m, respectively. Same for the others; ③GE stands for germination rate; GR stands for germination potential; GI stands for germination index.由图1可以看出,在不同电场强度和时间交互处理下的4个不同处理时间对发芽率影响不显著;发芽指数显示区域在t2-0.6和t3-0.8时呈现深红色,说明在此时间段下可以显著提高发芽指数;且在t1-0.2时显示区域呈现米白色,说明对发芽指数影响不大。发芽势在t3-0.8处理下显示区域呈现深红色,说明在此时间段下可以提高发芽势。综上,说明不同的电场强度和时间交互处理下对这3个指标的作用效果有所区别。电场处理下,根据卷荚相思种子各指标变化情况可将3个指标分为2类,指标发芽率单独聚为一类,发芽势和发芽指数聚为一类。
3. 讨论与结论
种子萌发期是植物生长发育初期阶段最为敏感的环节,关系到植物种子能否健康生长[19, 20]。发芽率、发芽势和发芽指数由种子最终发芽个数和萌发时间决定,可以反映种子萌发的生活力程度、发芽速率和整齐度[21]。本研究电场处理对卷荚相思种子萌发特性的影响与前人的研究结果基本一致,如武翠卿等[22]研究发现经高压静电场预处理后,谷子(Setaria italica)、荞麦(Fagopyrum esculentum Moench)及高粱(Sorghum bicolor)的千粒质量及产量都有增加,适宜的电场强度可促进谷子株高的增加,场强过低或者过高反而抑制株高的增加。李美清等[23]研究表明高压静电场处理改变了番茄(Solanum lycopersicum)的生长特性,适宜的电场对茎粗、鲜质量、叶绿素含量、根系形态具有促进作用,并提高产量。本研究发现不同电场强度和时间交互处理对卷荚相思种子的发芽率、发芽势和发芽指数具有不同程度的影响。发芽率在电场强度0.6 kV·cm−1、处理时间45 min时处于最佳状态,发芽势在电场强度0.8 kV·cm−1、处理时间45 min时均处于最高值,并且与CK有显著差异,发芽指数在电场强度0.6~0.8 kV·cm−1、处理时间30~45 min时均处于最优状态。说明卷荚相思种子在适宜的电场和时间交互处理对种子萌发有一定的作用,但是达到一定的电场阈值之后明显抑制种子的萌发。低压电场对种子萌发特性并没有促进作用,反而抑制了种子的萌发,这可能是卷荚相思种子表面附着一层保护膜,电场处理没有穿过种子表面的保护层,引起植物种子细胞内外物质的扩散[24]以及种子萌发反应所需要的养分条件。但高压电场对种子萌发特性具有抑制作用,这可能是电场回路里产生的电压太强直接击穿卷荚相思种子保护膜,对种子内部造成了一定的损伤,影响了植物种子体内电子传递和质子流的传递,并影响种子的萌发特性[25]。
植物种子的发芽率受本身特性和外界环境的双重影响,它们共同决定了植物的生长发育[26]。合理的电场强度和处理时间等对植物种子发芽率的提高具有重要作用[27]。前人研究表明,电场和时间两者交互处理下可以调控植物种子的发芽率[28]。在电场处理种子前,需要考虑电场强度的大小对植物种子的生物效应(在一定的场强范围内,才能引起生物效应),但是电场处理的作用时间对生物效应的影响也很重要,在选择最佳电场强度时也要考虑作用时间的长短,当电场强度和时间适宜时可以提升发芽速度并且提高发芽率[29]。本研究通过对不同电场强度和时间交互处理下对卷荚相思种子发芽率进行多元非线性回归分析,构建显著的多元非线性回归方程,得到不同电场强度和时间交互处理下最优非线性回归方程:Y=7.73+135.23X1+1.24X2−86.16X12−0.00911X22−0.75X1X2(R2=0.81,P<0.05)。非线性回归分析表明,卷荚相思种子发芽率与电场强度和处理时间之间存在显著(P<0.05)的相关性,且电场和时间两者交互处理能够影响卷荚相思种子发芽率。
通过聚类分析将发芽率、发芽势和发芽指数3个指标划分为2类,第1类为发芽率,第2类为发芽势和发芽指数组成。从分类结果上看,卷荚相思种子萌发期的发芽指数表现趋势较为显著。发芽指数放大种子活力特征,使活力不同的种子差异更加明显[30]。刘继宏[31]研究发现静电处理种子可以加速种子的新陈代谢,促进种子的萌发,并有效提高种子活力,增强酶活性。有研究发现电场的回路会产生电晕放电,可击穿空气中的NO和O3,与空气中的水发生反应,并使种子内部的生理条件发生变化,有利于种子的萌发[32, 33]。这可能是电场和时间交互处理下激发种子体内的潜力,改变种子膜内外电位差以及物质交换的速度,从而影响种子体内各种酶的活性,最终影响种子的萌发特性。
综上所述,卷荚相思种子发芽率在电场强度0.6 kV·cm−1、处理时间45 min时处于最高状态;发芽势和发芽指数均在电场强度0.8 kV·cm−1、处理时间45 min时处于最高状态。试验表明不同电场强度和时间两者交互处理对卷荚相思种子的萌发起到了促进作用但也产生了阻碍作用,造成不同的作用效果取决于电场强度和处理时间。结果表明适宜的电场强度和时间交互处理能够刺激卷荚相思种子的内部各种贬藏物质由休眠状态转变为活跃状态从而促进种子萌发,改善种子的活力。
-
图 9 DcbHLH14基因在低温(A)、干旱(B)和ABA(C)处理下的表达情况
*表示与0 h差异显著(P<0.05);***表示与0 h极显著差异(P<0.001);
Figure 9. Expressions of DcbHLH14 under cold stress (A), drought stress (B), and ABA treatment (C)
* represents significant difference from control (0 h) at P<0.05; *** represents extremely significant difference from control (0 h) at P<0.001.
图 10 DcbHLH14基因在低温、干旱和ABA处理下表达水平的qRT-PCR分析结果
不同小写字母表示同一处理不同时间之间差异显著(P<0.05)。
Figure 10. qRT-PCR analysis on expressions of DcbHLH14
under low temperature, drought, and ABA treatment Different lowercase letters indicant significant difference among different treatment times of the same treatment (P<0.05).
表 1 DcbHLH14基因启动子顺式作用元件分析结果
Table 1 Cis-acting elements in DcbHLH14 promoter
序号 No. 元件名称 Element 元件序列 Element sequence 数目 Number 功能预测 Predicted function 1 MYCCONSENSUSAT CANNTG 18 低温响应 Low temperature responsive 2 MYBCORE CNGTTR 4 水分胁迫调控 Regulation of water stress 3 MYB2CONSENSUSAT YAACKG 3 脱水响应 Dehydration responsive 4 LTRECOREATCOR15 CCGAC 3 低温响应 Low temperature responsive 5 MYBATRD22 CTAACCA 2 脱水响应 Dehydration responsive 6 MYB1AT WAACCA 1 脱水响应 Dehydration responsive 7 ABRELATERD1 ACGTG 1 ABA响应 Abscisic acid responsive -
[1] 蔡琳, 彭鹏. 名贵中药铁皮石斛化学成分及其药理作用浅述 [J]. 安徽化工, 2021, 47(1):24−25. DOI: 10.3969/j.issn.1008-553X.2021.01.008 CAI L, PENG P. A brief introduction on the chemical constituents and pharmacological action of rare Chinese medicine Dendrobium officinale [J]. Anhui Chemical Industry, 2021, 47(1): 24−25.(in Chinese) DOI: 10.3969/j.issn.1008-553X.2021.01.008
[2] 黄嘉雯, 陈小阳, 刘涛利, 等. 花色素苷合成关键调节基因的研究进展 [J]. 分子植物育种, 2019, 17(11):3602−3608. DOI: 10.13271/j.mpb.017.003602 HUANG J W, CHEN X Y, LIU T L, et al. Research progress of the key regulatory genes for anthocyanin synthesis [J]. Molecular Plant Breeding, 2019, 17(11): 3602−3608.(in Chinese) DOI: 10.13271/j.mpb.017.003602
[3] 王力伟, 房永雨, 刘红葵, 等. bHLH转录因子的研究进展 [J]. 畜牧与饲料科学, 2020, 41(1):23−27. DOI: 10.12160/j.issn.1672-5190.2020.01.005 WANG L W, FANG Y Y, LIU H K, et al. Research progress of bHLH transcription factors [J]. Animal Husbandry and Feed Science, 2020, 41(1): 23−27.(in Chinese) DOI: 10.12160/j.issn.1672-5190.2020.01.005
[4] 陈清, 汤浩茹, 董晓莉, 等. 植物Myb转录因子的研究进展 [J]. 基因组学与应用生物学, 2009, 28(2):365−372. CHEN Q, TANG H R, DONG X L, et al. Progress in the study of plant myb transcription factors [J]. Genomics and Applied Biology, 2009, 28(2): 365−372.(in Chinese)
[5] AMOUTZIAS G, VERON A, WEINER J, et al. One billion years of bZIP transcription factor evolution: Conservation and change in dimerization and DNA-binding site specificity [J]. Molecular Biology and Evolution, 2006, 24(3): 827−835. DOI: 10.1093/molbev/msl211
[6] 覃超. 甜瓜CmbHLH93和CmbHLH130基因在果实发育中的作用[D]. 呼和浩特: 内蒙古大学, 2020: 84-86. QIN C. The role of CmbHLH93 and CmbHLH130 in fruit development of melon[D]. Hohhot: Inner Mongolia University, 2020: 84-86. (in Chinese)
[7] 张娇. 两个bHLH转录因子(AtLPl和AtLP2)在拟南芥细胞伸长生长中的功能研究[D]. 武汉: 华中师范大学, 2019: 28-29. ZHANG J. Study on the roles of two bHLH transcription factors(AtLPl and AtLP2)in cell elongation of Arabidopsis thaliana[D]. Wuhan: Central China Normal University, 2019: 28-29. (in Chinese)
[8] LIU Y, LI X, LI K, et al. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis [J]. PLoS Genetics, 2013, 9(10): e1003861. DOI: 10.1371/journal.pgen.1003861
[9] 宋建辉. bHLH113调控拟南芥开花和花青素合成的分子机制研究[D]. 杭州: 浙江农林大学, 2020. SONG J H. The molecular regulatory mechanism of flowering and anthocyanin by bHLH113 in Arabidopsis[D]. Hangzhou: Zhejiang A & F University, 2020. (in Chinese)
[10] WU H H, REN Z Y, ZHENG L, et al. The bHLH transcription factor GhPAS1 mediates BR signaling to regulate plant development and architecture in cotton [J]. The Crop Journal, 2021, 9(5): 1049−1059. DOI: 10.1016/j.cj.2020.10.014
[11] ZHANG J H, LV H Z, LIU W J, et al. bHLH transcription factor SmbHLH92 negatively regulates biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza [J]. Chinese Herbal Medicines, 2020, 12(3): 237−246. DOI: 10.1016/j.chmed.2020.04.001
[12] MENG F W, YANG C, CAO J D, et al. A bHLH transcription activator regulates defense signaling by nucleo-cytosolic trafficking in rice [J]. Journal of Integrative Plant Biology, 2020, 62(10): 1552−1573. DOI: 10.1111/jipb.12922
[13] 尹航. 露地菊CgbHLH113基因的克隆及功能分析[D]. 哈尔滨: 东北林业大学, 2021. YIN H. Cloning and functional analysis of CgbHLH113 gene from Chrysanthemum × grandiflora[D]. Harbin: Northeast Forestry University, 2021. (in Chinese)
[14] LI Y Y, SUI X Y, YANG J S, et al. A novel bHLH transcription factor, NtbHLH1, modulates iron homeostasis in tobacco (Nicotiana tabacum L.) [J]. Biochemical and Biophysical Research Communications, 2020, 522(1): 233−239. DOI: 10.1016/j.bbrc.2019.11.063
[15] YI K K, WU Z C, ZHOU J, et al. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice [J]. Plant Physiology, 2005, 138(4): 2087−2096. DOI: 10.1104/pp.105.063115
[16] WANG F B, ZHU H, CHEN D H, et al. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana [J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2016, 125(2): 387−398. DOI: 10.1007/s11240-016-0953-1
[17] CHEN H C, CHENG W H, HONG C Y, et al. The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively [J]. Rice, 2018, 11(1): 50. DOI: 10.1186/s12284-018-0244-z
[18] GAO Y, WU M Q, ZHANG M J, et al. Roles of a maize phytochrome-interacting factors protein ZmPIF3 in regulation of drought stress responses by controlling stomatal closure in transgenic rice without yield penalty [J]. Plant Molecular Biology, 2018, 97(4): 311−323.
[19] REN Y R, YANG Y Y, ZHAO Q, et al. MdCIB1, an apple bHLH transcription factor, plays a positive regulator in response to drought stress [J]. Environmental and Experimental Botany, 2021, 188: 104523. DOI: 10.1016/j.envexpbot.2021.104523
[20] ZHAO Q, XIANG X H, LIU D, et al. Tobacco transcription factor NtbHLH123 confers tolerance to cold stress by regulating the NtCBF pathway and reactive oxygen species homeostasis [J]. Frontiers in Plant Science, 2018, 9: 381. DOI: 10.3389/fpls.2018.00381
[21] DONG H Z, CHEN Q M, DAI Y Q, et al. Genome-wide identification of PbrbHLH family genes, and expression analysis in response to drought and cold stresses in pear (Pyrus bretschneideri) [J]. BMC Plant Biology, 2021, 21(1): 86. DOI: 10.1186/s12870-021-02862-5
[22] JIN R, KIM H S, YU T, et al. Identification and function analysis of bHLH genes in response to cold stress in sweetpotato [J]. Plant Physiology and Biochemistry, 2021, 169: 224−235. DOI: 10.1016/j.plaphy.2021.11.027
[23] YU Z M, ZHANG G H, TEIXEIRA DA SILVA J A, et al. The methyl jasmonate-responsive transcription factor DobHLH4 promotes DoTPS10, which is involved in linalool biosynthesis in Dendrobium officinale during floral development [J]. Plant Science, 2021, 309: 110952. DOI: 10.1016/j.plantsci.2021.110952
[24] CHEN Y, WANG Y Z, LYU P, et al. Comparative transcriptomic analysis reveal the regulation mechanism underlying MeJA-induced accumulation of alkaloids in Dendrobium officinale [J]. Journal of Plant Research, 2019, 132(3): 419−429. DOI: 10.1007/s10265-019-01099-6
[25] 张志勇, 阳静, 齐泽民. 铁皮石斛总RNA提取方法的比较研究 [J]. 江苏农业科学, 2017, 45(4):33−35. DOI: 10.15889/j.issn.1002-1302.2017.04.009 ZHANG Z Y, YANG J, QI Z M. Comparative study on extraction methods of total RNA from Dendrobium candidum [J]. Jiangsu Agricultural Sciences, 2017, 45(4): 33−35.(in Chinese) DOI: 10.15889/j.issn.1002-1302.2017.04.009
[26] ZHANG G Q, LIU K W, LI Z, et al. The Apostasia genome and the evolution of orchids [J]. Nature, 2017, 549(7672): 379−383. DOI: 10.1038/nature23897
[27] WANG Y, LIU A Z. Genomic characterization and expression analysis of basic Helix-loop-Helix (bHLH) family genes in traditional Chinese herb Dendrobium officinale [J]. Plants (Basel, Switzerland), 2020, 9(8): 1044.
[28] 李季生, 李娜, 贾漫丽, 等. 基于转录组数据挖掘桑树bHLH转录因子家族 [J]. 分子植物育种, 2022, 20(6):1798−1810. DOI: 10.13271/j.mpb.020.001798 LI J S, LI N, JIA M L, et al. Mining bHLH transcription factor family of mulberry based on transcriptome data [J]. Molecular Plant Breeding, 2022, 20(6): 1798−1810.(in Chinese) DOI: 10.13271/j.mpb.020.001798
[29] 王菊萍, 王珍, 张铁军, 等. 蒺藜苜蓿MtbHLH148转录因子的克隆与转化及其功能分析 [J]. 西北植物学报, 2019, 39(6):963−973. DOI: 10.7606/j.issn.1000-4025.2019.06.0963 WANG J P, WANG Z, ZHANG T J, et al. Cloning and analysis of a basic Helix-loop-Helix (bHLH) transcription factor MtbHLH148 from Medicago truncatula L [J]. Acta Botanica Boreali-Occidentalia Sinica, 2019, 39(6): 963−973.(in Chinese) DOI: 10.7606/j.issn.1000-4025.2019.06.0963
[30] SUN W J, JIN X, MA Z T, et al. Basic helix-loop-helix (bHLH) gene family in Tartary buckwheat (Fagopyrum tataricum): Genome-wide identification, phylogeny, evolutionary expansion and expression analyses [J]. International Journal of Biological Macromolecules, 2020, 155: 1478−1490. DOI: 10.1016/j.ijbiomac.2019.11.126
[31] 杨贞, 蔡友铭, 张永春, 等. 基于SRAP分子标记的铁皮石斛遗传多样性分析 [J]. 上海农业学报, 2019, 35(5):23−27. DOI: 10.15955/j.issn1000-3924.2019.05.05 YANG Z, CAI Y M, ZHANG Y C, et al. Genetic diversity analysis of Dendrobium officinale based on SRAP molecular markers [J]. Acta Agriculturae Shanghai, 2019, 35(5): 23−27.(in Chinese) DOI: 10.15955/j.issn1000-3924.2019.05.05
[32] 朱璐璐, 周波. bHLH蛋白在植物发育及非生物胁迫中的调控[J/OL]. 分子植物育种, 2021: 1-14. (2021-02-23). https://kns.cnki.net/kcms/detail/46.1068.S.20210222.1744.012.html. ZHU L L, ZHOU B. Regulation of bHLH protein in plant development and abiotic stress[J/OL]. Molecular Plant Breeding, 2021: 1-14. (2021-02-23). https://kns.cnki.net/kcms/detail/46.1068.S.20210222.1744.012.html.(in Chinese)
[33] CASTILHOS G, LAZZAROTTO F, SPAGNOLO-FONINI L, et al. Possible roles of basic helix-loop-helix transcription factors in adaptation to drought [J]. Plant Science, 2014, 223: 1−7. DOI: 10.1016/j.plantsci.2014.02.010
[34] LIU Y J, JI X Y, NIE X G, et al. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs [J]. The New Phytologist, 2015, 207(3): 692−709. DOI: 10.1111/nph.13387
[35] JI X Y, NIE X G, LIU Y J, et al. A bHLH gene from Tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation [J]. Tree Physiology, 2016, 36(2): 193−207.
[36] 耿晶晶. 甜橙bHLH家族转录因子发掘及CsbHLH18抗寒功能鉴定与作用机制解析[D]. 武汉: 华中农业大学, 2018: 77-79. GENG J J. Genome-wide identification of bHLH transcription factor family in sweet orange(Citrus sinensis) and functional characterization and mechanism analysis of CsbHLH18 in cold resistance[D]. Wuhan: Huazhong Agricultural University, 2018: 77-79. (in Chinese)
[37] PARK S, LEE C, DOHERTY C J, et al. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network [J]. The Plant Journal, 2015, 82(2): 193−207. DOI: 10.1111/tpj.12796
[38] CHINNUSAMY V, ZHU J K, SUNKAR R. Gene regulation during cold stress acclimation in plants [J]. Methods in Molecular Biology (Clifton, N J), 2010, 639: 39−55.
[39] CHINNUSAMY V, OHTA M, KANRAR S, et al. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis [J]. Genes & Development, 2003, 17(8): 1043−1054.
[40] SHI Y T, DING Y L, YANG S H. Molecular regulation of CBF signaling in cold acclimation [J]. Trends in Plant Science, 2018, 23(7): 623−637. DOI: 10.1016/j.tplants.2018.04.002




 下载:
下载: