Genetic Evolution of F1L, B2L, and GIF of Orf Virus in Four Clinical Samples
-
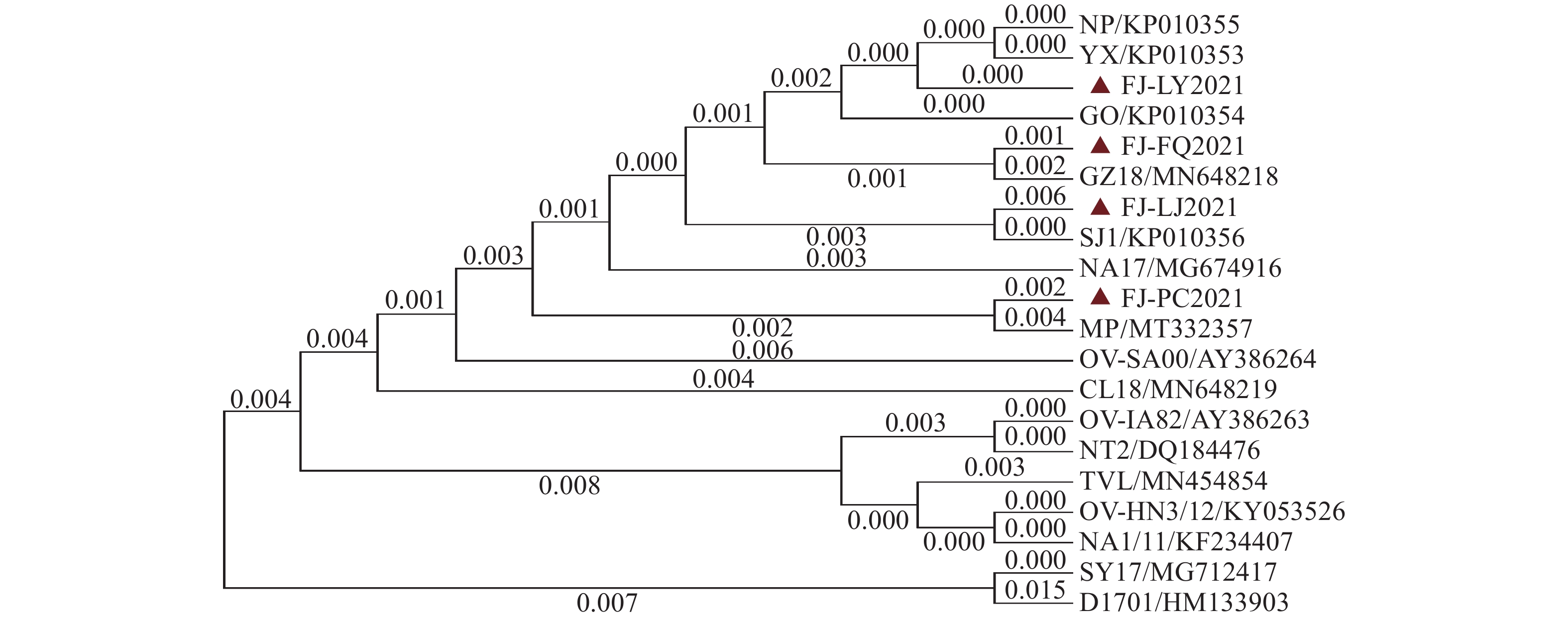
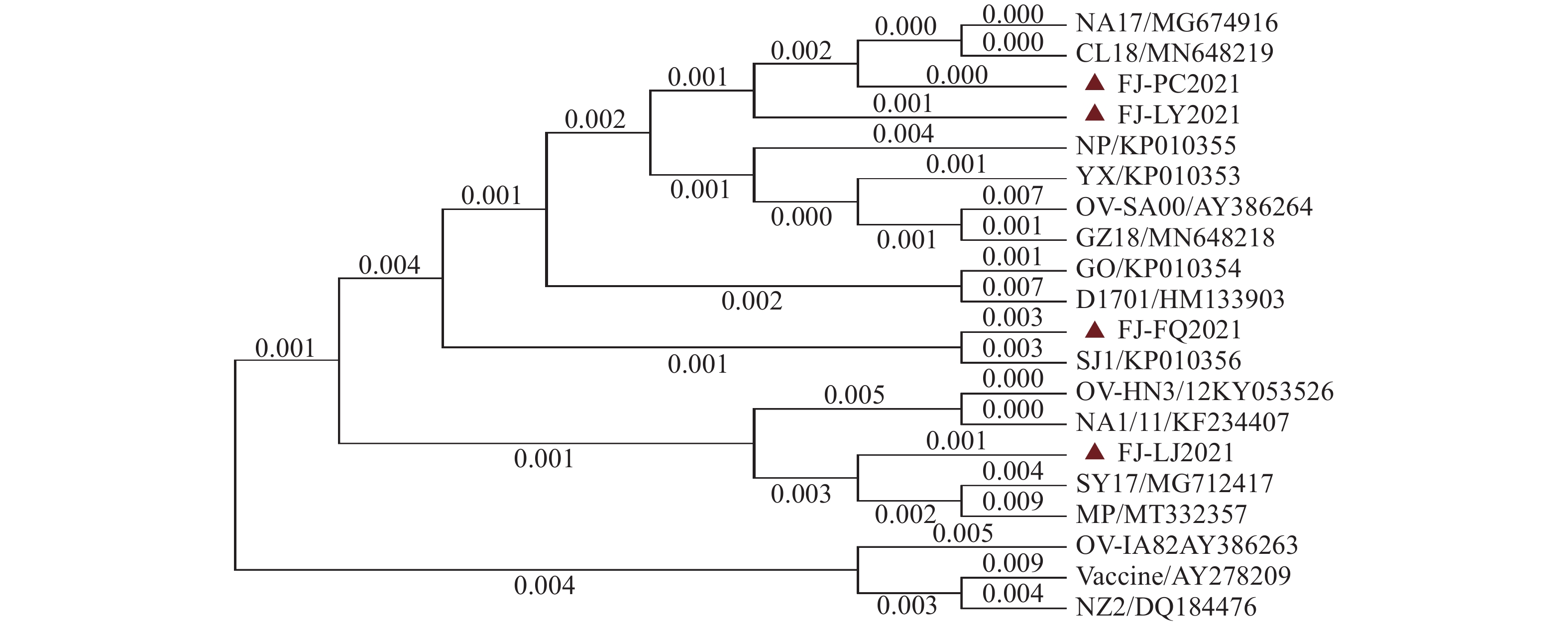
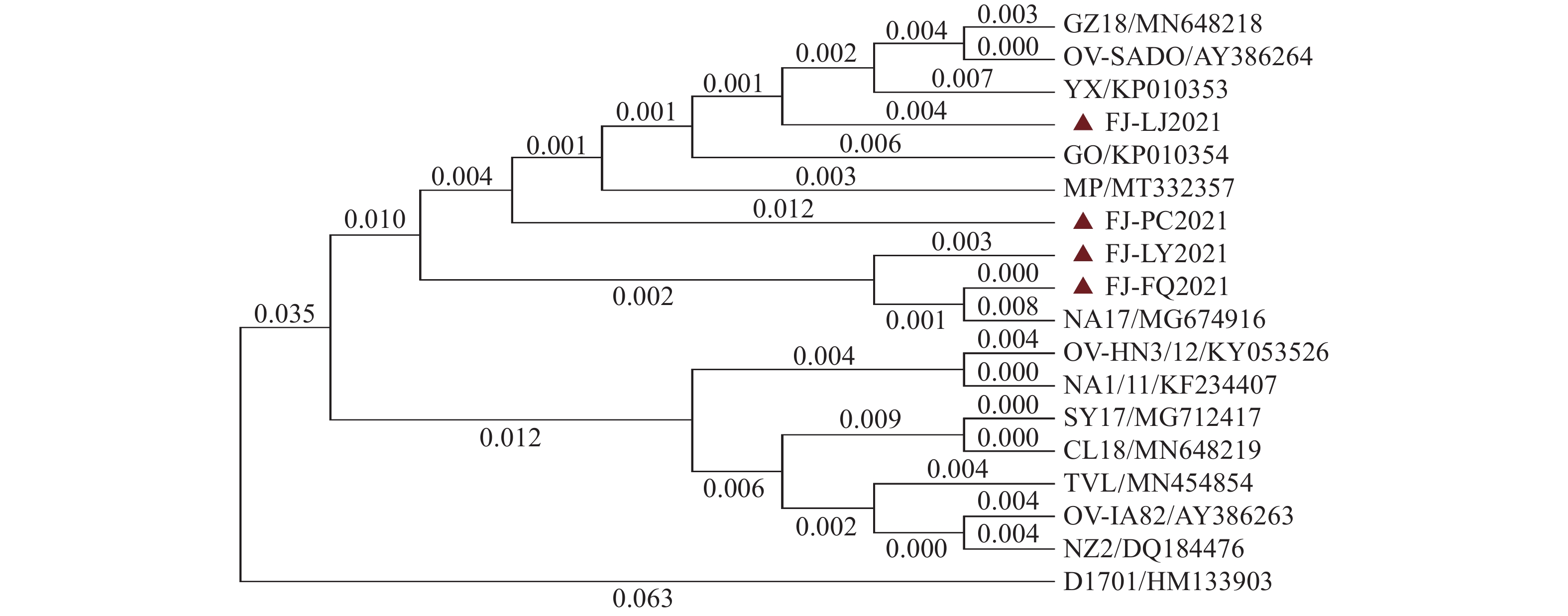
摘要:目的 研究2021年福建省部分羊场羊口疮病毒(Orf virus, ORFV)分子流行病学情况。方法 对2021年采集的4份病料经PCR鉴定ORFV阳性的临床样品分别克隆其F1L、B2L和GIF基因,将获得的克隆质粒送往测序公司进行测序,将测序拼接后的序列上传至GenBank数据库中,利用生物信息学软件进行对比分析。结果 4份临床样品中ORFV的F1L、B2L和GIF基因核苷酸序列相似性分别为98.0%~98.8%、98.4%~99.5%和97.2%~99.6%;与国内流行毒株核苷酸序列相似性分别为97.4%~99.4%、97.4%~99.9%和95.7%~99.0%;与国外流行毒株核苷酸序列相似性分别为97.5%~98.6%、97.9%~98.9%和95.6%~98.0%;与德国D1701弱毒株核苷酸序列相似性分别为96.5%~96.8%、98.0%~98.7%和96.0%~96.4%,与NZ2参考株核苷酸序列相似性分别为97.7%~98.1%、97.5%~98.1%和95.9%~96.1%。遗传进化树结果显示4份临床样品中ORFV与吉林(NA17、SY17和CL18株)、福建SJ1株和印度MP株亲缘关系较近;与美国OV-IA82株、德国D1701弱毒株、新西兰NZ2参考株和中国疫苗株亲缘关系较远;表明当前流行毒株可能是本省毒株、国内吉林GZ18株和国外印度MP株的重组毒株。结论 获得4份临床样品中ORFV B2L、F1L和GIF基因序列并上传至GenBank数据库中,丰富了ORFV相关基因序列的数据库,同时阐明福建省部分羊场当前ORFV的流行特征,为福建省今后有效防控Orf提供理论依据。Abstract:Objective Molecular epidemiology of the Orf virus (ORFV) in the diseased sheep and goats from farms in Fujian Province in 2021 were studied.Method The ORFV genes, F1L, B2L, and GIF, from 4 clinical samples of the diseased animals were confirmed positive by PCR prior to cloning for sequence determination. The data were uploaded to GenBank for a comparative analysis using bioinformatics software.Result The nucleotide sequence similarities were 98.0%–98.8% on F1L , 98.4%–99.5% on B2L, and 97.2%–99.6% on GIF; those related to the domestic epidemic strains, 97.4%–99.4% on F1L, 97.4%–99.9% on B2L, and 95.7%–99.0% on G1F; those related to the foreign epidemic strains, 97.5%–98.6% on F1L, 97.9%–98.9% on B2L, and 95.6%–98.0% on G1F; those related to the German D1701 attenuated strain, 96.5%–96.8% on F1L, 98.0%–98.7% on B2L, and 96.0%–96.4% on G1F; and those related to the NZ2 reference strain, 97.7%–98.1% on F1L, 97.5%–98.1% on B2L, and 95.9%–96.1% on G1F. The phylogenetic tree showed the ORFVs closely related to the strains from Jilin (NA17, SY17, and CL18), Fujian (SJ1), and India (MP), but remotely with OV-IA82 from the USA, D1701 attenuated strain from German, NZ2 reference strain from New Zealand, and the vaccine strain from China. It appeared that the current epidemic strain might be a recombination of Fujian, Jilin GZ18, and Indian MP strains.Conclusion The sequences of B2L, F1L, and GIF from the 2021 clinical specimens were uploaded to GenBank database enriching its collection. This study characterized the ORFVs found in the sheep and goats infected by the epidemic and facilitated further studies in the disease control and prevention.
-
Keywords:
- Orf virus /
- F1L /
- B2L /
- GIF /
- evolution analysis
-
0. 引言
【研究意义】基因组作为特定生物全部染色体的遗传物质的总和,能反映物种全部和特定的遗传信息[1],其大小(C值)指单倍体细胞中全套染色体的DNA总量,基因组大小的度量一般以重量计算,以质量 (Picogram,pg)或碱基对数目(Million base pair,Mb)为单位,1 pg约等于978 Mb[2-3]。C值的测定不但能估测物种的DNA含量,还能为目标物种后续基因组测序、基因组学及进化生物学研究提供数据参考,除此之外也是生物学家进行物种分类和种群进化的重要依据[4-6]。朱砂根(A. crenata)隶属紫金牛科(Myrsinaceae)紫金牛属(Ardisia),是一种兼具药用和观赏价值的重要经济植物,朱砂根在分子生物学和基因组学层面的相关研究则刚刚起步,通过测定朱砂根种质资源的基因组大小,对后续开展朱砂根基因组学及其他相关研究具有重要意义。【前人研究进展】目前,植物DNA C值数据库(Plant DNA C-values Database;https://cvalues.science.kew.org/)的数据,已收录了12 273个物种的C值数据,其中包括10 770种被子植物。近年来朱砂根植物研究内容主要集中于药用价值[7-8]、栽培繁殖[9-11]、生理生化[12-13]以及遗传多样性[14-15]等方面,目前朱砂根在分子生物学和基因组学层面的研究主要集中在叶绿体基因组和特定功能基因等领域,例如刘雄伟等[16]通过对朱砂根和其变种红凉伞的叶绿体全基因组进行测序,解析两者的叶绿体基因组特征及差异,从而确定朱砂根在紫金牛属系统发育位置;刘畅等[17]通过对朱砂根AcGGPPS基因进行克隆测序研究了其生物学特性,为今后该基因功能的研究奠定基础,杨君[18]通过转录组测序,对朱砂根三萜皂苷合成的相关基因进行差异分析,发现了93个差异表达基因直接影响朱砂根三萜皂苷含量。【本研究切入点】有关朱砂根基因组大小测定方面的研究鲜见报道,一定程度上限制了朱砂根分子生物学及基因组学等研究领域进一步进展。【拟解决的关键问题】本研究以番茄(Lycopersicon esculentum)为内标,建立基于流式细胞术测定朱砂根基因组大小的测定方法和流程,估算24份朱砂根资源的基因组大小,以期为今后朱砂根基因组文库的建立、基因组全序列测定及其基因组学研究等工作的开展提供基础数据。
1. 材料与方法
1.1 试验材料
24份朱砂根材料见(表1),其中22份朱砂根是通过自然变异并经过多年人工选育,最终形成表型性状稳定遗传的栽培品种,6个获得新品种权,16个实地审查;而2份野生朱砂根种质由课题组骆亮等从武夷山收集并种植于福建农林大学森林兰苑苗圃。内参样本来自中国科学院昆明植物研究所,选取番茄种子萌发后1月的嫩叶作为试验材料。24份朱砂根试验材料的嫩叶取自福建农林大学苗圃中心,在中国西南野生生物种质资源库分子生物学实验平台对样品进行流式细胞仪检测。
表 1 24份朱砂根供试材料采样信息Table 1. Sampling of 24 A. crenata germplasms编号 No. 种质名称 Germplasm name 种质类别 Germplasm type 编号 No. 种质名称 Germplasm name 种质类别 Germplasm type Z-01 赤丹 Chi Dan 栽培品种 Cultivated variety Z-13 福株 Fu Zhu 栽培品种 Cultivated variety Z-02 锦绣 Jin Xiu 栽培品种 Cultivated variety Z-14 金边富贵 Jin Bian Fu Gui 栽培品种 Cultivated variety Z-03 绿翡翠 Green Jade 栽培品种 Cultivated variety Z-15 金玉满堂 Jin Yu Man Tang 栽培品种 Cultivated variety Z-04 粉佳人 Pink Beauty 栽培品种 Cultivated variety Z-16 竹叶富贵 Bamboo Leaf Fu Gui 栽培品种 Cultivated variety Z-05 平安富贵 Ping An Fu Gui 栽培品种 Cultivated variety Z-17 玛瑙红 Onyx Red 栽培品种 Cultivated variety Z-06 金冠 Golden Crown 栽培品种 Cultivated variety Z-18 碧珠 Bi Zhu 栽培品种 Cultivated variety Z-07 福满堂 Fu Man Tang 栽培品种 Cultivated variety Z-19 龙珠 Long Zhu 栽培品种 Cultivated variety Z-08 碧霞珠 Bixia Zhu 栽培品种 Cultivated variety Z-20 霞珠 Xia Zhu 栽培品种 Cultivated variety Z-09 金富贵 Jin Fu Gui 栽培品种 Cultivated variety Z-21 仙桃 Xian Tao 栽培品种 Cultivated variety Z-10 梁野富贵 Liang Ye Fu Gui 栽培品种 Cultivated variety Z-22 珠塔 Zhu Ta 栽培品种 Cultivated variety Z-11 赤玲珑 Red Lingerie 栽培品种 Cultivated variety Z-(A) 朱砂根A A. crenata A 野生种质 Wild germplasm Z-12 大富贵 Da Fu Gui 栽培品种 Cultivated variety Z-(B) 朱砂根B A. crenata B 野生种质 Wild germplasm 1.2 试验方法
1.2.1 细胞悬浮液制备
解离液的选择对于试验结果的准确性至关重要。本试验解离液选择的是mGb[45 mmol·L−1 MgCl2·6H2O,20 mmol·L−1 MOPS,30 mmol·L−1柠檬酸钠,1%(m/V)PVP-40,0.2%(V/V)TritonX-100,10 mmol·L−1 Na2EDTA,20 μL·mL−1β-巯基乙醇,pH 7.5[19]。
将样品置于0.8 mL预冷的mGb解离液中,用锋利的刀片将组织迅速垂直切碎,使其在解离液中冰上静置10 min,然后用40 μm孔径滤网过滤,即得到细胞核悬浮液。在细胞核悬液加入适当体积预冷的碘化丙啶(Propidium iodide,PI)(母液浓度1 mg·mL−1)和适当体积的RNAase溶液(母液浓度1 mg·mL−1),置于冰上避光染色0.5~1.0 h,并控制所有待测样品染色时间一致。PI染液和RNAase溶液的工作浓度均为50 μg·mL−1 [19-21]。
1.2.2 流式细胞仪检测
以番茄为内参,将待测样品的悬液和内参样品的悬液按适当比例混合,利用BD FACScalibur流式细胞仪对染色后的细胞核悬浮液样品上机检测。采用488 nm蓝光激发,检测PI的发射光荧光强度,每次检测收集10000个颗粒。变异系数(CV)控制在5%以内。
1.2.3 基因组大小计算
PI在着色过程中的嵌入量与DNA量呈正比关系,故荧光强度可以表示出基因组DNA的相对含量。观察待测样品和对照植物PI-DNA复合体的荧光峰值,即可得出2种植物DNA含量的比值,再乘以内参植物的C值,即可计算出待测植物的C值。计算公式如下:
待测样品DNA含量=内参DNA含量×待测样品的荧光强度/内参样品的荧光强度[19]。
1.3 数据分析
利用Excel2010、Modifit3.0分析软件作图分析。
2. 结果与分析
本研究采用流式细胞术对24份朱砂根的染色体DNA总量进行检测。对比番茄和24样品基因组单独测定的结果可见,番茄基因组的测定峰与朱砂根基因组测定峰两者无重叠(图1),具有良好的区分度,证明使用番茄作为内参样本具备可靠性。
选择的内参样本番茄基因组大小为900 Mbp,根据测定结果分析计算,得到了24份朱砂根样品的基因组大小(表2)。结果发现,大部分朱砂根种质资源的C值在1.8 Gb左右,24份朱砂根种质C值平均为1.87 Gb。栽培品种中,玛瑙红(Z-17)、霞珠(Z-20)、珠塔(Z-22)C值最小,均为1.77 Gb,赤丹C值最大,达到2.41 Gb,共有9个朱砂根品种的基因组大小均超过了2 Gb,高于平均C值;收集的2份野生朱砂根种质基因组大小别为1.79、1.80 Gb,接近平均C值。从24份基因组大小检测结果来看,朱砂根种内C值存在明显变异;同时朱砂根种质资源从野生到人工选育栽培过程中发生了基因组大小变异。
表 2 流式细胞术测定的24份朱砂根品种资源基因组大小Table 2. Genome sizes of 24 A. crenata determined by flow cytometry样品编号Sample No. 内参荧光强度Internal reference fluorescence intensity 待测样品荧光强度Fluorescence intensity of the sample to be measured 比值Ratio 基因组大小Genome/Gb 样品编号Sample No. 内参荧光强度Internal reference fluorescence intensity 待测样品荧光强度Fluorescence intensity of the sample to be measured 比值Ratio 基因组大小Genome/Gb Z-01 18.90 51.79 2.74 2.41 Z-13 26.61 63.08 2.37 2.09 Z-02 23.83 48.40 2.03 1.79 Z-14 27.09 65.13 2.40 2.12 Z-03 24.33 49.58 2.04 1.79 Z-15 27.22 64.87 2.38 2.10 Z-04 26.06 55.25 2.12 1.87 Z-16 26.68 55.70 2.09 1.84 Z-05 18.75 48.60 2.59 2.28 Z-17 26.63 53.53 2.01 1.77 Z-06 21.30 52.18 2.45 2.16 Z-18 27.19 55.46 2.04 1.79 Z-07 22.72 56.47 2.49 2.19 Z-19 27.61 55.73 2.02 1.78 Z-08 22.44 55.92 2.49 2.19 Z-20 28.26 56.70 2.01 1.77 Z-09 24.16 51.55 2.13 1.88 Z-21 28.56 58.22 2.04 1.79 Z-10 26.36 55.40 2.10 1.85 Z-22 28.34 56.89 2.01 1.77 Z-11 26.07 59.64 2.29 2.01 Z-(A) 28.77 58.62 2.04 1.79 Z-12 25.35 51.83 2.04 1.80 Z-(B) 28.54 58.29 2.04 1.80 3. 讨论
流式细胞术作为一种广泛应用于动植物生命科学各个领域的单细胞分选技术,融合了激光技术、半导体技术、流体力学、细胞化学等多学科知识,相较于孚耳根微显影法、基因组测序法更简便易行,检测结果更稳定可靠,已逐渐成为动植物基因组大小测定、倍性鉴定等研究的重要工具[22-24];并且流式细胞术可通过内参和外参法2种方式对物种进行核DNA含量检测,其中内参法可避免因样品的差异、机器的不稳定等因素造成的误差[25]。本研究首次以番茄作为内参样本对朱砂根种质资源基因组大小进行了测定,结果在所有混合组织匀浆处理中,DNA含量在上机检测时控制的变异系数CV均处于5%以内,内参样本和待测样本峰不重合且无杂峰,表明本次试验结果稳定可靠[26];排除了试验本身造成朱砂根种质资源基因组大小变异这一因素。
通常认为每个生物体的基因组大小是恒定数值,与生物体的进化水平及生物体复杂程度均无关,即所谓“C值悖论”[27];随后有学者认为基因组大小与植物体结构和功能的复杂程度呈正相关关系,但后期研究结果证明没有正相关关系[28]。如今随着基因组大小进化相关研究的不断深入,越来越多的研究表明物种C值与表型特征、核型特征、种子特征以及生态环境因子等众多因素密切相关[5,29-30]。而真核生物基因组长度的巨大差异主要归因于重复序列,其中种内不同种群间的基因组长度变异主要是由卫星DNA和转座子造成[31]。本次试验检测计算得到24份朱砂根基因组大小为1.77~2.41 Gb,平均为1.87 Gb,测定结果发现,部分朱砂根种质间C值存在明显变异,野生资源与部分栽培品种间也存在明显变异;结合24朱砂根实地观察以及前人研究,发现朱砂根品种间表型遗传丰富[14],居群间基因流动较密切且存在较高的遗传渗透[15]。因此,笔者推测朱砂根种内基因组大小变异的主要原因可能与其丰富的遗传变异或分子突变而导致种内重复序列的变化[32-33]。另外,有研究发现生物基因组大小的变异还与其多倍化现象有关[34]。但目前关于朱砂根种质资源倍性研究尚不清楚,因此不排除朱砂根中存在染色体非整倍化或多倍化现象,从而导致其种内基因组大小变异[32]。总之,影响朱砂根种内基因组大小因素很多,具体影响机制还需进一步研究。
4. 结论
本研究首次利用流式细胞技术对朱砂根基因组大小的测定方法进行了初步探索,并成功估测了24份朱砂根种质资源的基因组大小,丰富了朱砂根基因组基本信息。同时,通过测定朱砂根核DNA的含量,不仅为朱砂根的全基因组测序工作的前期预算评估和后期组装结果矫正提供数据指导,同时也为后续朱砂根基因组学和进化生物学研究提供参考。
-
表 1 引物序列
Table 1 Sequences of primers
引物名称
Primers引物序列
Sequences(5'-3')退火温度
Annealing/℃片段大小
Product size/bpF1L-F CGGAATTCGATGGATCCACCCGAAATCACG 55 1029 F1L-R CCGTCGAGTCACACGATGGCCGTGACCA B2L-F CGGAATTCGATGTGGCCGTTCTCCTCCATC 55 1137 B2L-R CCGTCGAGTTAATTTATTGGCTTGCAGAACTCC GIF-F CGGAATTCGATGGCGTGCCTCAGAGTGTT 52 798 GIF-R CCGTCGAGTTAGCGCGCCGTCTGCATGC 表 2 ORFV 参考毒株
Table 2 Reference ORFV strains
毒株
Virus登录号
Locus分离地
Isolate时间
TimeNZ2
OV-SA00
OV-IA82
D1701
GZ18
SJ1
NP
NA17
YX
GO
OV-HN3/12
NA1/11
MP
SY17
CL18
TVL
China vaccineDQ184476
AY386364
AY386363
HM133903
MN648218
KP010356
KP010355
MG674916
KP010353
KP010354
KY053526
KF234407
MT332357
MG712417
MN648219
MN454854
JQ904789New Zealand
USA
USA
Germany
Jilin, China
Fujian, China
Fujian, China
Jilin, China
Fujian, China
Fujian, China
Guangzhou, China
Guangzhou, China
India
Jilin, China
Jilin, China
West Texas, USA
Gansu, China2005
2003
2003
2010
2018
2015
2011
2016
2012
2012
2011
2013
2017
2016
2018
2019
2012 -
[1] CORADDUZZA E, SANNA D, ROCCHIGIANi A M, et al. Molecular insights into the genetic variability of Orf virus in a mediterranean region (Sardinia, Italy) [J]. Life (Basel), 2021, 11(5): 416.
[2] KHALAFALLA A I, ELHAG A E, ISHAG H Z A. Field investigation and phylogenetic characterization of orf virus (ORFV) circulating in small ruminants and Pseudocowpoxvirus (PCPV) in dromedary camels of eastern Sudan [J]. Heliyon, 2020, 6(3): e03595. DOI: 10.1016/j.heliyon.2020.e03595
[3] TOBLER C, RITTER-SCHENK C, ZIMMERMANN P. Orf virus infection: Ecthyma contagiosum [J]. The Journal of Pediatrics, 2022, 243: 236−237. DOI: 10.1016/j.jpeds.2021.11.067
[4] HOSAMANI M, SCAGLIARINI A, BHANUPRAKASH V, et al. Orf: an update on current research and future perspectives [J]. Expert Review of Anti-Infective Therapy, 2009, 7(7): 879−893. DOI: 10.1586/eri.09.64
[5] ALAJLAN A M, ALSUBEEH N A. Orf (ecthyma contagiosum) transmitted from a camel to a human: A case report [J]. The American Journal of Case Reports, 2020, 21: e927579.
[6] COSTA R A, CARGNELUTTI J F, SCHILD C O, et al. Outbreak of contagious ecthyma caused by Orf virus (Parapoxvirus ovis) in a vaccinated sheep flock in Uruguay [J]. Brazilian Journal of Microbiology, 2019, 50(2): 565−569. DOI: 10.1007/s42770-019-00057-7
[7] ALDAIF B A, MERCER A A, FLEMING S B. The Parapoxvirus Orf virus inhibits IFN-β expression induced by dsRNA [J]. Virus Research, 2022, 307: 198619. DOI: 10.1016/j.virusres.2021.198619
[8] BUKAR A M, JESSE F F A, ABDULLAH C A C, et al. Immunomodulatory strategies for parapoxvirus: current status and future approaches for the development of vaccines against orf virus infection [J]. Vaccines (Basel), 2021, 9(11): 1341. DOI: 10.3390/vaccines9111341
[9] GELAYE E, ACHENBACH J E, JENBERIE S, et al. Molecular characterization of orf virus from sheep and goats in Ethiopia, 2008-2013 [J]. Virology Journal, 2016, 13: 34. DOI: 10.1186/s12985-016-0489-3
[10] SCAGLIARINI A, GALLINA L, DAL P F, et al. Heparin binding activity of orf virus F1L protein [J]. Virus Research, 2004, 105(2): 107−112. DOI: 10.1016/j.virusres.2004.04.018
[11] OEM J K, ROH I S, LEE K H, et al. Phylogenetic analysis and characterization of Korean orf virus from dairy goats: Case report [J]. Virology Journal, 2009, 6: 167. DOI: 10.1186/1743-422X-6-167
[12] MARTINS M, RODRIGUES F S, JOSHI L R, et al. Orf virus ORFV112, ORFV117 and ORFV127 contribute to ORFV IA82 virulence in sheep [J]. Veterinary Microbiology, 2021, 257: 109066. DOI: 10.1016/j.vetmic.2021.109066
[13] CHEN H, LI W, KUANG Z, et al. The whole genomic analysis of orf virus strain HN3/12 isolated from Henan province, central China [J]. BMC veterinary research, 2017, 13(1): 260. DOI: 10.1186/s12917-017-1178-1
[14] 林裕胜, 江锦秀, 江斌, 等. 福建省羊传染性脓疱病毒F1L、B2L和VIR基因的遗传变异分析 [J]. 动物医学进展, 2017, 38(2):1−6. DOI: 10.3969/j.issn.1007-5038.2017.02.001 LIN Y S, JIANG J X, JIANG B, et al. Genetic variation analysis of F1L, B2L and VIR genes of orf virus in Fujian Province [J]. Progress in Veterinary Medicine, 2017, 38(2): 1−6.(in Chinese) DOI: 10.3969/j.issn.1007-5038.2017.02.001
[15] 张家确, 吴景央. 一起人畜共患羊传染性脓疱病的调查 [J]. 中国兽医杂志, 2006, 42(9):53−54. DOI: 10.3969/j.issn.0529-6005.2006.09.035 ZHANG J Q, WU J Y. Investigation of a zoonotic infectious orf disease [J]. Chinese Journal of Veterinary Medicine, 2006, 42(9): 53−54.(in Chinese) DOI: 10.3969/j.issn.0529-6005.2006.09.035
[16] WANG Y, YANG K, WANG Y A, et al. Identification and phylogenetic analysis of an orf virus strain isolated in Anhui Province, East-central China, in 2018 [J]. Acta Virologica, 2019, 63(3): 270−277. DOI: 10.4149/av_2019_304
[17] AHANGER S A, PARVEEN R, NAZKI S, et al. Detection and phylogenetic analysis of Orf virus in Kashmir Himalayas [J]. VirusDisease, 2018, 29(3): 405−410. DOI: 10.1007/s13337-018-0473-1
-
期刊类型引用(2)
1. 李玉洁. 南繁育种中种质资源基因漂移的知识产权问题与法律法规保护对策. 分子植物育种. 2024(13): 4500-4504 .  百度学术
百度学术
2. 周子琦,孙楠,严雅洁,杨志远,李良俊,冯凯. 慈姑基因组大小鉴定及核型分析. 西北植物学报. 2024(07): 1122-1128 .  百度学术
百度学术
其他类型引用(0)




 下载:
下载: