Polymorphisms and Expressions of FABP3 in Tibetan and Yorkshire Pigs
-
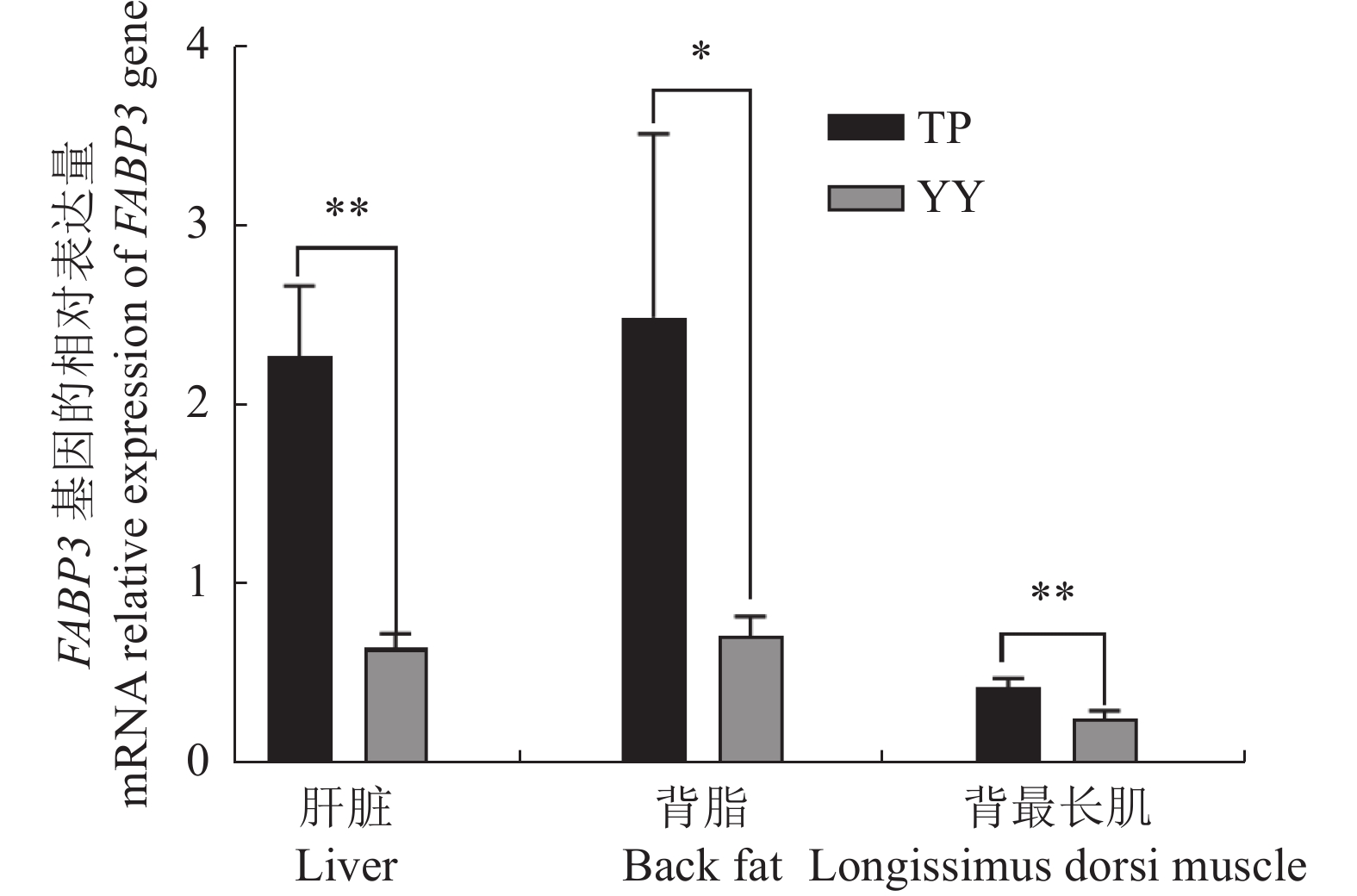
摘要:目的 脂肪酸结合蛋白3(FABP3)参与长链脂肪酸的摄取及利用,在脂肪沉积中发挥重要作用。探究FABP3基因在藏猪和大约克猪间的基因多态性和表达差异可为藏猪品质改良提供分子机制参考。方法 选取180日龄藏猪和大约克猪为研究对象,对两猪种FABP3基因5'侧翼区和CDS区进行单核苷酸多态性(SNPs)筛选,使用实时荧光定量检测FABP3基因在肝脏、背最长肌和背脂3个组织中的表达量。结果 在FABP3基因5'侧翼区筛选到T-114C和C-635A等2个SNPs位点,且SNPs位点基因型频率在藏猪与大约克猪群体间均呈极显著差异(P < 0.01);经转录因子预测发现这2个SNPs位点与前脂肪细胞的更新、分化以及脂肪沉积相关,推测这两个多态性位点是参与调控FABP3基因表达的重要功能位点。FABP3基因在藏猪肝脏、背最长肌中的表达量极显著高于大约克猪(P<0.01),在背脂中的表达量显著高于大约克猪(P < 0.05)。结论 推测FABP3基因可能为调控藏猪脂肪代谢的重要候选基因,在藏猪脂肪沉积中呈正向调控作用。Abstract:Objective Fatty acid binding protein 3 (FABP3) is involved in the uptake and utilization of long-chain fatty acids and plays an important role in fat deposition. To investigate the gene polymorphism and expression differences of FABP3 gene between Tibetan and York pigs could help improve the quality of Tibetan pigs for the genetic level.Methods In this study, single nucleotide polymorphisms (SNPs) in the 5' flanking region and CDS region of FABP3 in randomly selected 180-d-old Tibetan and Yorkshire pigs were tested. Expressions of FABP3 in the liver, longest dorsal muscle, and dorsal fat were detected using real-time fluorescence.Results Two SNPs, T-114C and C-635A, were found in FABP3 with significantly differentiated genotype frequencies between the two species of pigs (P<0.01). Upon transcription factor prediction, these 2 SNPs loci were found to be associated with preadipocyte renewal, differentiation, and fat deposition, and it was hypothesized that they were important functional loci involved in the regulation of FABP3 gene expression. The expressions of FABP3 in the liver and longest dorsal muscle of Tibetan pigs were extremely significantly higher than those of Yorkshire pigs (P<0.01), and that in the dorsal fat significantly higher than that of Yorkshire pigs (P<0.05).Conclusion FABP3 might be closely related to the regulation of fat metabolism and deposition of Tibetan pigs which differs from Yorkshire variety.
-
Keywords:
- Tibetan pig /
- FABP3 /
- fat deposition /
- polymorphic loci
-
0. 引言
【研究意义】紫薇(Lagerstroemia indica)为千屈菜科紫薇属灌木或小乔木,大多为灌木,花量大,花色艳丽,花期长,在园林中常作夏季观花树种应用。全世界紫薇属植物约有55种,主要分布在亚洲东部、东南部、南部和澳大利亚的北部[1],中国是紫薇属植物的世界分布中心和起源中心[2],共有24种[3],其中福建紫薇(L. limii)是我国特有植物,常为小乔木,基部少分枝,是改良紫薇株型的优异种质[4],但与紫薇相比,其花色仅有粉色和紫色,花期短(单株花期一般20余天),利用福建紫薇和紫薇杂交,以期获得花色美丽、花期长、植株体量大的乔木型紫薇新品种。
【前人研究进展】株型是农作物、园艺作物和观赏植物最重要的性状之一。一方面,它限定了植物本身各器官的空间排列,与产量、品质及植株外观有着紧密联系[5−7];另一方面,株型显著地影响了群体结构、田间小气候及群落结构,进而影响光能利用率、作物产量及景观效果[8,9]。株型主要由株高、分枝等性状共同决定。紫薇株型有低矮紧凑型、匍匐型(垂枝型)、普通灌木型、小乔木型等类型[10]。课题组前期用紫薇与乔木型的大花紫薇(L. speciosa)杂交获得种间杂交品种‘风华绝代’ [11],从紫薇品种‘Pocomoke’开放授粉群体中筛选出垂枝品种‘玲珑’,用屋久岛紫薇和‘Pocomoke’杂交培育出地被型品种‘绿地毯’,用屋久岛紫薇(L. fauriei)和‘Pocomoke’杂交后回交得到低矮紧凑型品种‘粉精灵’[12],这些品种极大丰富了紫薇的株型。
株型性状包括株高、节间长度、节间数量、分枝数量、分枝角度等,研究株型性状的遗传规律及形成机制,对定向培育株型各异的植物(尤其是木本植物)新品种具有重要的指导意义[13]。刘阳[14]以屋久岛紫薇为母本,‘Pocomoke’为父本构建了杂交群体,后代的株型性状有明显的分离,推断株高性状可能由主效基因控制,微效基因修饰。石俊[15]在屋久岛紫薇בCreole’杂交群体中发现直立型:中间型:匍匐型符合1∶2∶1分离比,推测紫薇匍匐性状受一对不完全显性的主效基因控制,受微效基因修饰。焦垚[16]用大花紫薇与‘Dynamite'的正、反交群体发现株高、冠幅、当年生枝条长度、当年生枝条粗度等8个性状出现广泛分离,各性状之间呈极显著正相关。
【本研究切入点】目前,园林中栽植的小乔木型或乔木型紫薇多是紫薇古树或大树,其乔木型主要依靠持续的修剪维持,仍带有较强的灌木生长性状。园林中的单干紫薇高度通常仅约1米,尚未达到小乔木高度的标准要求[17]。观赏性状好的乔木型紫薇品种比较缺乏。培育花色多样、乔木型的紫薇品种成为紫薇育种的一个重要方向。【拟解决的关键问题】因此,本研究以福建紫薇和开花繁密、花期长的紫薇品种‘千层绯雪’为亲本进行正反交,利用主基因+多基因混合遗传模型[18−20]对一年生杂交后代的株型性状进行分析,以期为乔木型紫薇品种选育提供依据。
1. 材料与方法
1.1 试验材料
以小乔木型的福建紫薇(FJ)和花量大的灌木型紫薇品种‘千层绯雪’(QC)为亲本进行杂交,杂交在北京林业大学国家花卉工程技术研究中心小汤山基地进行。
1.2 方法
2021年6月参考刘阳[14]的方法进行杂交,以‘千层绯雪’为母本的组合作为正交。10月底至11月初采收果实,翌年3月种子播种于72孔穴盘(泥炭∶蛭石=1∶1)中,待幼苗长出第二对真叶后移栽至口径12 cm的花盆(泥炭∶蛭石=3∶1)中;5月份将杂交后代带花盆移至室外(昼温18 ℃~38 ℃;夜温12 ℃~28 ℃);11月份再移回温室(5 ℃~20 ℃),期间正常水肥管理。2022年12月份测定正、反交群体的株高(基质表面到植株顶端的距离)、地径(仅有一个主枝的后代,测量基质表面处直径;基部有多个分枝的,选最粗的枝条测量基部直径)、一级分枝点高度(主枝第一次分枝处距基质表面的高度)、一级分枝数(主枝上的分枝数量)、植株开张角度(测量侧枝与主枝之间两个垂直方向的角度,取平均值)、侧枝GSA(侧枝与垂直方向的夹角)[15]、节间长度(第3~5叶位相邻两个芽点之间的距离)。
1.3 数据分析
用EXCEL软件整理数据,参考焦垚[16]的方法,对杂交后代性状进行描述性统计并计算变异系数(coefficient of variation,CV);对7个株型性状进行主成分分析与相关性分析;根据双假测交策略,将正、反交群体视为用于遗传分析的假F2群体,参考王靖天[21]的方法,用R语言SEA包(Segregation Analysis)分析性状的最适遗传模型,SEA2.0软件包由南京农业大学章元明教授提供。
2. 结果与分析
2.1 杂交座果率
福建紫薇与‘千层绯雪’之间杂交亲和性好,座果率高(75.5%以上),果实可自然发育成熟,种子播种后正常发芽(表1)。
表 1 福建紫薇与‘千层绯雪’的杂交结实率Table 1. seed setting rate of L.limii and L. indica ‘Qianceng Feixue’母本×父本
Maternal×
Paternal杂交数量
Flower Number
of crosses结实数
Fruit number结实率
Fruiting
rate/%出苗数
Seedling
number‘千层绯雪’×
福建紫薇371 280 75.5 963 福建紫薇×
‘千层绯雪’17 17 100.0 66 合计 388 297 76.5 1029 2.2 正、反交群体的株型性状
QC×FJ杂交组合共获得963株后代,一年生株高从1.5 cm到24.5 cm,其中1.5~5 cm(不包括5 cm)的后代占32.37%,5~15 cm的占64.69%,15 cm以上(包括15 cm)占3.84%;地径从0.30 mm到4.44 mm,其中0.3~0.8 mm(不包括0.8 mm)占14.12%,0.8~2.2 cm占比75.50%,2.2cm以上(包括2.2 cm)占比10.38%;两参数的变异系数分别为51.97%、43.57%,变异丰富。具有分枝的后代有171株,占17.76%,一级分枝点高度从0.8 cm到6.2 cm,分枝数量在0~4个之间,变异系数为240.42%。963株后代中,株高在15 cm以上、地径2.2 mm以上,没有分枝的后代有23株,占比2.39%。
株高、地径、开张角度、侧枝GSA基本呈正态分布(图2),偏度均为正值,分别是1.32、1.09、1.89、0.71(表2),表示直方图向正态分布区域的左侧倾斜,4个性状的峰度分别为2.16、1.55、6.43、0.05,表示正态分布陡峭。
表 2 QC×FJ群体的描述性统计Table 2. Plant type difference in hybrid population性状
Traits最大值
Maximum最小值
Minimum均值
Average标准差
SD偏度
Skewness峰度
Kurtosis变异系数
CV/%株高
Plant height/cm24.5 1.50 6.87 357 1.32 2.16 51.97 地径
Diameter/mm4.44 0.30 1.40 0.61 1.09 1.55 43.57 一级分枝点高度
Height at first branching point/cm6.20 0.80 3.31 1.05 0.11 0.19 31.72 一级分枝数
Number of first-order branches/each4 0 0.24 0.577 2.591 6.839 240.42 开张角度
Opening angle/°87.56 8.20 27.90 11.35 1.89 6.43 40.68 侧枝GSA
Lateral branch GSA/°72.25 15.35 35.26 12.42 0.71 0.05 35.22 节间长度
Internode length/cm2.0 0.4 0.73 0.26 1.58 3.23 35.62 FJ×QC组合共获得66株杂交后代,该群体一年生株高从2.0 cm到22.0 cm,其中株高2~5 cm的后代占30.30%,5~15 cm的占62.12%,15 cm以上的占7.58%;地径从0.44 mm到2.98 mm,其中0.44~0.8 mm的后代占13.64%,0.8~2.2 cm的占72.72%,2.2 cm以上的占13.64%。两者的变异系数分别为54.11%、42.86%,变异丰富。66株后代中,具有分枝的后代有15株,占22.73%,一级分枝点高度从0.5 cm到5.8 cm,分枝数量在0~2个之间,变异系数为196.15%。该群体中,株高15 cm以上、地径2.2 mm以上、没有分枝的后代有1株,占1.52%。
6个性状的频率分布直方图见图2,地径、开张角度基本呈正态分布(图3),偏度均为正值,分别为0.91、0.99(表3),表示直方图向正态分布区域的左侧倾斜;这2个性状的峰度分别为0.15、0.97,表示正态分布陡峭。
表 3 FJ×QC群体的描述性统计Table 3. Descriptive statistics of the FJ×QC population性状
Traits最大值
Maximum最小值
Minimum均值
Average标准差
SD偏度
Skewness峰度
Kurtosis变异系数
CV/%株高
Plant height/cm22.0 2.0 7.54 4.08 1.06 2.19 54.11 地径
Diameter/mm2.98 0.44 1.40 0.60 0.91 0.15 42.86 一级分枝点高度
Height at first branching point/cm5.8 0.5 2.41 1.39 0.99 0.97 57.68 一级分枝数
Number of first-order branches/each2 0 0.26 0.51 1.84 2.69 196.15 开张角度
Opening angle/°59.69 3.03 27.34 19.70 −1.38 1.121 72.06 侧枝GSA
Lateral branch GSA/°48.35 10.9 24.65 11.33 0.83 −0.10 45.96 节间长度
Internode length/cm54.8 17.3 33.40 13.39 0.38 −1.05 40.08 2.3 主成分分析与性状相关性
对QC×FJ与FJ×QC群体进行相关性分析(表6,表7),相同点是两个群体株高与地径、一级分枝数与开张角度、开张角度与侧枝GSA均呈极显著正相关,株高与节间长度极显著或显著正相关。说明植株越高,地径越粗,节间越长,生长越快;开张角度越大,一级分枝数越多,侧枝GSA越大。
表 6 不同遗传模型下QC×FJ 群体的 AIC 值Table 6. AIC values of QC×FJ population under different genetic models模型 Model implication 株高
Plant height地径
Diameter开张角度
Opening angle侧枝GSA
Lateral branch GSA一级分枝点高度
Height at first branching pointA-0 0MG 5188.884 391.072 1303.792 1334.032 506.3505 A-1 1MG-AD 4899.472 300.2864 1250.446 1313.571 503.0956 A-2 1MG-A 4942.795 306.6786 1248.605 1312.763 501.3233 A-3 1MG-EAD 5032.944 335.7514 1282.295 1318.325 506.3529 A-4 1MG-NCD 4993.272 331.1677 1287.728 1321.534 509.4304 B-1 2MG-ADI 4950.868 325.7444 1278.015 1314.864 515.8107 B-2 2MG-AD 4871.614 294.8911 1245.498 1305.003 497.1486 B-3 2MG-A 4910.321 302.0399 1249.31 1314.201 510.264 B-4 2MG-EA 4888.363 295.9231 1246.738 1303.926 491.0738 B-5 2MG-CD 5192.889 395.0742 1307.791 1338.033 510.3601 B-6 2MG-EAD 5190.889 393.0743 1305.791 1336.033 508.3601 加粗值代表最小 AIC 值;MG 代表主基因模型;A 代表加性效应;D 代表显性效应; I 代表上位性 效应;E 代表相等;N 代表负向;C 代表完全。
The values in bold indicate the minimum AIC value; MG represents the major gene model; A represents the additive effect; D represents the dominance effect; I represents epistatic interaction;E represents equal; N represents negative; C represents complete. The same as below.表 7 不同遗传模型下FJ×QC群体的 AIC 值Table 7. AIC values of FJ×QC population under different genetic models模型
ModelModel implication 地径
Diameter开张角度
Opening angleA-0 0MG 123.9251 118.3907 A-1 1MG-AD 109.1332 118.0483 A-2 1MG-A 114.9762 118.7385 A-3 1MG-EAD 115.9589 119.0364 A-4 1MG-NCD 110.7568 119.0415 B-1 2MG-ADI 110.3914 122.8118 B-2 2MG-AD 100.7582 106.6611 B-3 2MG-A 111.5877 118.2622 B-4 2MG-EA 107.1847 112.1628 B-5 2MG-CD 127.9204 122.358 B-6 2MG-EAD 125.9204 120.358 不同的地方是QC×FJ群体株高与一级分枝点高度、开张角度、侧枝GSA存在极显著正相关性,说明植株越高,一级分枝点越高、开张角度与侧枝GSA也越大,这些参数在FJ×QC中无相关性。FJ×QC群体一级分枝点高度与一级分枝数呈极显著正相关,而在QC×FJ中呈不显著的负相关。
2.4 混合遗传模型分析
QC×FJ群体中株高、地径、开张角、侧枝GSA、一级分枝点高度五个性状基本呈正态分布,我们对这些性状进行了混合遗传分析。一级分枝数、节间长度两个性状不符合正态分布,无法进行分析。FJ×QC群体可用于分析的株型性状有地径、开张角度。
不同遗传模型的AIC值见表8和9,从中选择AIC值最小或相对较小的模型作为备选最适模型再进行适合性检验(表10)。根据结果,正、反交群体的株高、地径、开张角的最适模型是2MG-AD,由2对加性-显性主基因控制;侧枝GSA、一级分枝点高度的最适模型是2MG-EA,由2对等加性主基因控制(表11)。QC×FJ群体株高、地径、开张角的| da | 均大于 | db |,说明第一对主基因的加性效应更显著,三个性状的显性效应ha、hb均为负值,起减效作用。FJ×QC群体地径与开张角两个性状的| da | 均大于 | db |,说明第一对主基因的加性效应更显著,ha、hb均为负值,起减效作用。
表 8 QC×FJ、FJ×QC 群体入选模型的适合性检验Table 8. Fitness test for QC×FJ and FJ×QC population inclusion models群体
Group性状
Traits模型
ModelU12 U22 U32 nW2 Dn QC×FJ 株高
Plant height2MG-AD 0.0251 (0.8741 )0.0453 (0.8314 )0.0565 (0.8121 )0.0393 (0.9369 )0.0206 (0.7996 )地径
Diameter2MG-AD 0.0004 (0.9851 )0.0005 (0.9822 )0.0003 (0.9866 )0.012(1) 0.0107 (0.9998 )开张角度
Opening angle2MG-AD 0.0015 (0.9692 )0.0004 (0.9927 )0.0127 (0.9103 )0.0077 (1.0008 )0.0184 (1)侧枝GSA
Lateral branch GSA2MG-EA 0.0008 (0.9776 )0.0013 (0.971)0.0647 (0.7992 )0.0206 (0.9964 )0.032(0.993) 一级分枝点高度
Height at first branching point2MG-EA 0.0014 (0.9704 )0.001( 0.9748 )0.0003 (0.986)0.064( 0.7929 )0.0621 (0.5055 )FJ×QC 地径
Diameter2MG-AD 0.0013 (0.9708 )0.0017 (0.9674 )0.0005 (0.9828 )0.0063 (1.0046 )0.0267 (1)开张角度
Opening angle2MG-AD 0.0217 (0.883)0.0142 (0.9051 )0.0087 (0.9257 )0.0152 (0.9996 )0.1078 (0.9871 )U12, U22 和 U32 为均匀性检验,nW2 为 Smirnov 检验,Dn 为 Kolmogorov 检验,括号内为概率值。
U12, U22and U32 represent the uniformity tests; nW2 represents the Smirnov test; and Dn represents the Kolmogorov test, values in brackets are probability.表 9 QC×FJ、FJ×QC(一年生)群体各性状在最适模型下的遗传参数Table 9. Genetic parameters of traits in QC×FJ and FJ×QC population under optimal models群体
Group性状
Traitsda(d) db ha(h) hb i jab jba l h2mg /% QC×FJ 株高
Plant height3.5662 1.5173 −1.452 −0.782 0 地径
Diameter0.3965 0.1399 −0.226 −0.29 0 开张角度
Opening angle8.4046 6.8204 −3.912 −3.831 0 侧枝GSA
Lateral branch GSA8.8543 23.802 一级分枝点高度
Height at first branching point0.6373 63.933 FJ×QC 地径
Diameter0.6005 0.3401 −0.238 −0.257 91.1643 开张角度
Opening angle10.415 7.135 −1.855 −3.439 95.6964 da:第 1 对主基因加性效应;db:第 2 对主基因加性效应;ha:第 1 对主基因显性效应;hb:第 2 对主基因显性效应;i:加×加效应;jab:加×显应;jba:显×加效应;l:显×显效应;h2mg:主基因遗传率。
da: the first major-gene additive effect; db: the second major-gene additive effect; ha: the first major-gene dominant effect; hb: the second major-gene dominant effect; i: additivity × additivity effect; jab: additivity × dominance effect; jba: dominance × additivity effect; l: dominance × dominance` effect; h2mg: indicates major gene heritability.3. 讨论
乔木一般具有明显的主干,株型高大,生长量大,植株基部无萌蘖枝或者较少。福建紫薇具有明显的乔木性状,与紫薇杂交亲和性高,是培育乔木型紫薇品种的优良亲本。株型性状变异的多样性是株型育种的基础,了解杂种后代性状的变异程度,对种质创新具有重要意义[22]。本研究获得的杂交群体的株高、地径、一级分枝点高度、一级分枝数等性状均存在不同程度的分离(图1),结合两个群体的变异系数,两群体的一级分枝数均存在显著变异。FJ×QC群体的一级分枝点高度、开张角度、侧枝GSA的变异系数比QC×FJ群体更大,说明福建紫薇作为母本时杂交后代有更丰富的株型变异。FJ×QC群体比QC×FJ群体杂交后代更倾向表现出植株高、地径粗、分枝数量多。因此,福建紫薇是培育生长量大、主干明显的紫薇品种的优良材料,与紫薇正反交后代均具选出生长迅速、株型高大的乔木型紫薇品种的可能。
幼苗期对株型提前进行选择,能缩短育种时间,加快育种进程[23]。焦垚[16]发现大花紫薇与紫薇的杂交后代中,花序的大小、花量、株型相关性状可以在早期通过叶片大小与植株体量进行选择。说明幼苗期对性状进行预选具有一定的可靠性。本研究对QC×FJ与FJ×QC正反交群体进行了相关性分析,发现两个群体均表现为,植株越高,地径越粗,节间越长,FJ×QC还表现为植株越高,一级分枝点越高。因此,为选育乔木型紫薇品种,可将植株高、地径粗、节间长、一级分枝点高度作为评价指标。在木本植物中,开花的起始是划分幼年期和成年期的标准[24]。乔木的幼年期与成年期会有形态、内源激素、分子调控上的差异[25]。因此,在幼年期筛选乔木性状仍存在一定的不确定性,需要后续进一步评估。
植物大多数表型性状都是由微效多基因控制的数量性状,在同一杂交后代群体的不同个体中往往表现出连续的数量差异,易受环境影响,需要应用统计学方法对整个群体进行测定和分析[26]。主基因+多基因遗传模型可以根据杂交后代的某个性状的表型观测值,推测出该性状的基因组分[27]。张琳[28]利用主基因+多基因混合遗传模型对牡丹杂交后代20个表型性状进行了分析,发现株高、冠幅等8个性状由微效多基因控制,新枝长度、花朵数等12个性状受一对或两对主基因控制。本研究对福建紫薇与‘千层绯雪’正、反交群体的株型性状进行了最适遗传模型分析,发现紫薇株高、地径的最适遗传模型为2MG-AD,开张角度、主枝GSA、一级分枝数、一级分枝点高度均受两对主基因控制,主基因间加性效应较稳定,对培育乔木型紫薇新品种具有重要的指导意义。但单世代的分离群体分析不能检测到多基因的存在,最适遗传模型尚有待进一步研究。后续可以构建多世代群体,来分析各株型性状的微效基因效应。
-
图 3 组织总RNA琼脂糖凝胶电泳
M:Marker;1~3分别为藏猪肝脏、背脂、背最长肌组织;4~6分别为大约克猪肝脏、背脂、背最长肌组织。
Figure 3. Agarose gel electrophoresis of total RNA in tissue
M:Marker; 1–3: Liver, dorsal fat, and longest dorsal muscle tissues of Tibetan pig, respectively; 4–6: liver, dorsal fat, and longest dorsal muscle tissues of Yorkshire pig, respectively.
图 4 FABP3基因在藏猪、大约克猪肝脏、背脂和背最长肌中的mRNA相对表达量
TP为藏猪,YY为大约克猪;*为显著差异(P < 0.05),**为极显著差异(P < 0.01)。
Figure 4. Relative expressions of FABP3 in liver, back fat, and longissimus dorsi muscle of TP and YY
TP: Tibetan pig; YY: Yorkshire pig; *: significant difference at P<0.05;**: extremely significant difference at P<0.01.
表 1 FABP3基因5'侧翼区和CDS区引物序列
Table 1 Primer sequence of 5' flanking region and CDS region in FABP3
引物
Primer扩增区域
Amplified region引物序列(5′-3′)
Primer sequences退火温度
Annealing temperature/℃产物大小
Product length/bp5′−FABP3-1 356 bp至−460 bp F: TCAGCCCAAGAGTGAGTTTC
R: CCTTCTTCCTCGAAAGCG56 817 5′−FABP3-2 −430 bp至−1365 bp F: TCTGCTGGCTCAAGTTCAGT
R: GAGAGGAGAAAGGAAACTCACT58 953 5′−FABP3-3 −1342 bp至−2197 bp F: TAGGAGTCAACTTTGGTGAGC
R: CCAACTGAACTTGAGCCAGCA59 856 5′−FABP3-4 −2195 bp至−3033 bp F: CTGGGAACCTCCATATGTCG
R: CTAAGCCACAATCTATCACCT57 849 FABP3-CDS 34 bp至445 bp F:CCTGTTCTGTCGTCTCTTTCTCA
R:TGCCTCTTTCTCGTAAGTGCG60 440 表 2 FABP3基因定量PCR引物序列
Table 2 Primer sequence of FABP3 for quantitative PCR
基因名称
Gene name登录号
GeneBank number引物序列(5′-3′)
Primer sequences退火温度
Annealing temperature/℃产物大小
Product length/bpFABP3 NM_001099931.1 F: ATGACCAAGCCTACCACAA
R: AAGTTTGCCTCCATCCAGT57 171 GAPDH NM_001206359.1 F: CACCATCTTCCAGGAGCGAG
R: CCCTTCAAGTGAGCCCCG57 120 表 3 FABP3基因SNPs位点基因型频率及卡方检验
Table 3 Genotype frequency and chi-square test of SNPs on FABP3
位点
Loci品种
Species样本量
Sample size基因型频率(个体数/频率)
Genotype frequency (Individuals/Frequency)基因频率
Gene frequencyχ2值
CardinalityP值
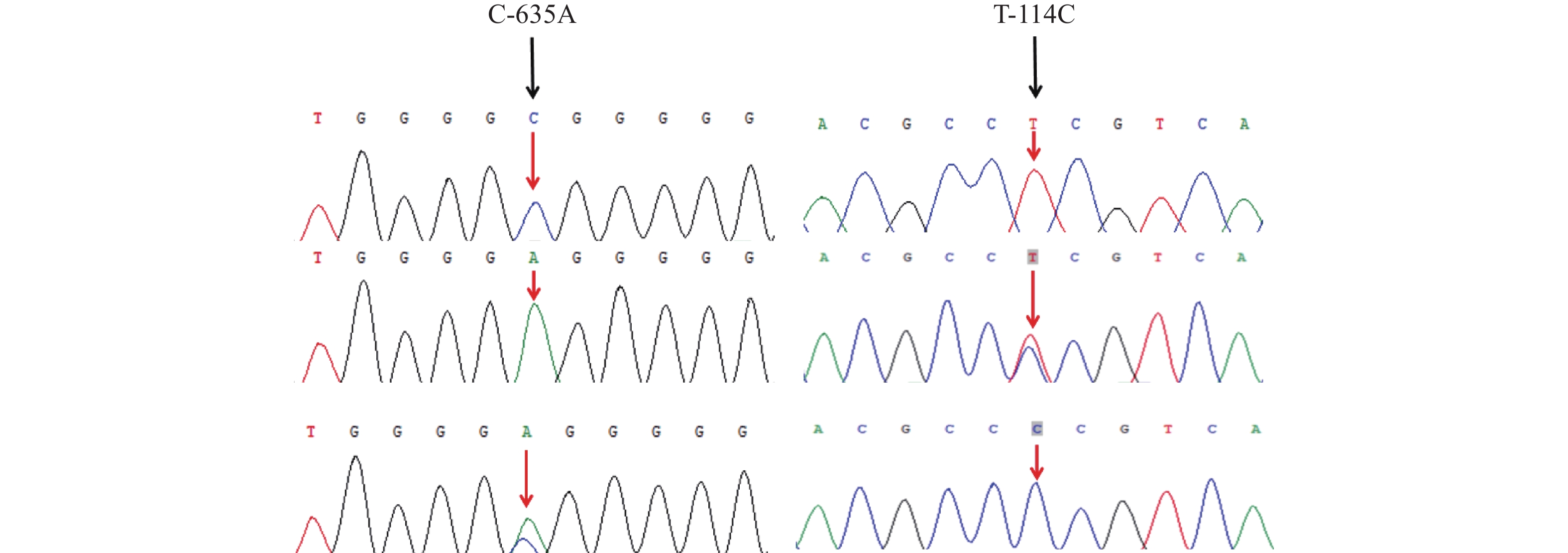
P valueCC CA AA C A C-635A 大约克猪 39 16/0.410 16/0.410 7/0.179 0.615 0.385 0.693 0.707 藏猪 28 15/0.520 12/0.410 28/1.000 0.000 1.000 0.000 1.000 藏猪 vs 大约克猪 χ2=43.979;P<0.01 T-114C TT TC CC T C 大约克猪 39 33/0.846 6/0.154 0/0 0.923 0.077 0.271 0.873 藏猪 28 0/0.000 1/0.036 27/0.964 0.018 0.982 0.009 0.995 藏猪 vs 大约克猪 χ2=63.476;P<0.01 表 4 FABP3基因SNPs位点转录因子预测结果
Table 4 Predicted transcription factors in FABP3 SNPs
突变位点
Mutation Loci突变前序列
Pre-mutation sequence突变后序列
Post-mutation sequence消失转录因子
Disappearance of transcription factors新增转录因子
Addition of transcription factorsC-635A TGGGGCGGGGG TGGGGAGGGGG ARNT2、 SUMO2、 RB1、 CTCF、SMAD4、LF11、CHD1、SUZ12、YY1 STAT5B、TCF7L2、TCF12、SREBP1、MYH11、SPI1、TP53、TBX21、HOXA9 T-114C ACGCCTCGTCA ACGCCCCGTCA E4F1 HES5、CLOCK、WT1、EP300、THAP11、KLF5 -
[1] SCHUMACHER M, DELCURTO-WYFFELS H, THOMSON J, et al. Fat deposition and fat effects on meat quality-a review [J]. Animals:an Open Access Journal from MDPI, 2022, 12(12): 1550.
[2] VITALI M, DIMAURO C, SIRRI R, et al. Effect of dietary polyunsaturated fatty acid and antioxidant supplementation on the transcriptional level of genes involved in lipid and energy metabolism in swine [J]. PLoS One, 2018, 13(10): e0204869. DOI: 10.1371/journal.pone.0204869
[3] KIM B, MIN Y J, JEONG Y, et al. Comparison of growth performance and related gene expression of muscle and fat from Landrace, Yorkshire, and Duroc and Woori black pigs [J]. Journal of Animal Science and Technology, 2023, 65(1): 160−174. DOI: 10.5187/jast.2022.e93
[4] LI Q G, TAO Z, SHI L H, et al. Expression and genome polymorphism of ACSL1 gene in different pig breeds [J]. Molecular Biology Reports, 2012, 39(9): 8787−8792. DOI: 10.1007/s11033-012-1741-6
[5] GRZES M, SADKOWSKI S, RZEWUSKA K, et al. Pig fatness in relation to FASN and INSIG2 genes polymorphism and their transcript level [J]. Molecular Biology Reports, 2016, 43(5): 381−389. DOI: 10.1007/s11033-016-3969-z
[6] LI A N, WU L J, WANG X Y, et al. Tissue expression analysis, cloning and characterization of the 5’-regulatory region of the bovine FABP3 gene [J]. Molecular Biology Reports, 2016, 43(9): 991−998. DOI: 10.1007/s11033-016-4026-7
[7] CHMURZYŃSKA A. The multigene family of fatty acid-binding proteins (FABPs): Function, structure and polymorphism [J]. Journal of Applied Genetics, 2006, 47(1): 39−48. DOI: 10.1007/BF03194597
[8] SWEENEY T, O'HALLORAN A M, HAMILL R M, et al. Novel variation in the FABP3 promoter and its association with fatness traits in pigs [J]. Meat Science, 2015, 100: 32−40. DOI: 10.1016/j.meatsci.2014.09.014
[9] HONG J, KIM D, CHO K, et al. Effects of genetic variants for the swine FABP3, HMGA1, MC4R, IGF2, and FABP4 genes on fatty acid composition [J]. Meat Science, 2015, 110: 46−51. DOI: 10.1016/j.meatsci.2015.06.011
[10] WANG L J, LI L, JIANG J, et al. Molecular characterization and different expression patterns of the FABP gene family during goat skeletal muscle development [J]. Molecular Biology Reports, 2015, 42(1): 201−207. DOI: 10.1007/s11033-014-3759-4
[11] LI B, ZERBY H N, LEE K. Heart fatty acid binding protein is upregulated during porcine adipocyte development [J]. Journal of Animal Science, 2007, 85(7): 1651−1659. DOI: 10.2527/jas.2006-755
[12] GERBENS F, VAN ERP A J, HARDERS F L, et al. Effect of genetic variants of the heart fatty acid-binding protein gene on intramuscular fat and performance traits in pigs [J]. Journal of Animal Science, 1999, 77(4): 846−852. DOI: 10.2527/1999.774846x
[13] SERÃO N V L, VERONEZE R, RIBEIRO A M F, et al. Candidate gene expression and intramuscular fat content in pigs [J]. Journal of Animal Breeding and Genetics, 2011, 128(1): 28−34. DOI: 10.1111/j.1439-0388.2010.00887.x
[14] 张敏. FABP3对LPS诱导的奶牛乳腺上皮细胞炎症反应的调控及机制研究[D]. 武汉: 华中农业大学, 2018. ZHANG M. Effects and mechanism of FABP3 gene on LPS-induced inflammation response in bovine mammary epithelial cells[D]. Wuhan: Huazhong Agricultural University, 2018. (in Chinese)
[15] JIANG Y, LIU J L, LIU H T, et al. miR-381-3p inhibits intramuscular fat deposition through targeting FABP3 by ceRNA regulatory network [J]. Biology, 2022, 11(10): 1497. DOI: 10.3390/biology11101497
[16] CHMURZYNSKA A, SZYDLOWSKI M, STACHOWIAK M, et al. Association of a new SNP in promoter region of the porcine FABP3 gene with fatness traits in a Polish synthetic line [J]. Animal Biotechnology, 2007, 18(1): 37−44. DOI: 10.1080/10495390600671560
[17] BLECHA I M Z, SIQUEIRA F, FERREIRA A B R, et al. Identification and evaluation of polymorphisms in FABP3 and FABP4 in beef cattle [J]. Genetics and Molecular Research:GMR, 2015, 14(4): 16353−16363. DOI: 10.4238/2015.December.9.3
[18] LI X P, KIM S W, CHOI J S, et al. Investigation of porcine FABP3 and LEPR gene polymorphisms and mRNA expression for variation in intramuscular fat content [J]. Molecular Biology Reports, 2010, 37(8): 3931−3939. DOI: 10.1007/s11033-010-0050-1
[19] WANG B B, LI P H, ZHOU W D, et al. Association of twelve candidate gene polymorphisms with the intramuscular fat content and average backfat thickness of Chinese suhuai pigs [J]. Animals:an Open Access Journal from MDPI, 2019, 9(11): 858.
[20] 强巴央宗, 张浩, 纪素玲, 等. 藏猪屠宰性能和肉质测定与分析 [J]. 中国畜牧杂志, 2008, 44(21):10−11,48. QIANG B, ZHANG H, JI S L, et al. Determination and analysis of slaughter performance and meat quality of Tibetan pigs [J]. Chinese Journal of Animal Science, 2008, 44(21): 10−11,48.(in Chinese)
[21] 商鹏, 强巴央宗, 张博, 等. 藏猪选育群屠宰性能和肉质测定分析 [J]. 黑龙江畜牧兽医, 2015(2):30−32. DOI: 10.13881/j.cnki.hljxmsy.2015.0092 SHANG P, Qiangbayangzong, ZHANG B, et al. Analysis of the slaughter performance and determination of meat quality in the selective breeding population of Tibetan pigs [J]. Heilongjiang Animal Science and Veterinary Medicine, 2015(2): 30−32.(in Chinese) DOI: 10.13881/j.cnki.hljxmsy.2015.0092
[22] 张一君. 微卫星DNA标记与SNP芯片鉴定宁乡猪亲缘关系[D]. 长沙: 湖南农业大学, 2017. ZHANG Y J. Phylogenetic analysis of Ningxiang pig based on microsatellite DNA markers and SNP chip[D]. Changsha: Hunan Agricultural University, 2017. (in Chinese)
[23] 尹杭. 猪BMP7和BMP15基因3’-UTR多态性及其与繁殖性能的关系[D]. 南京: 南京农业大学, 2020. YIN H. Polymorphism of the 3’-UTR of the porcine BMP7 and BMP15 and their association with reproductive performance[D]. Nanjing: Nanjing Agricultural University, 2020. (in Chinese)
[24] ARZATE-MEJÍA R G, RECILLAS-TARGA F, CORCES V G. Developing in 3D: The role of CTCF in cell differentiation [J]. Development, 2018, 145(6): 137729. DOI: 10.1242/dev.137729
[25] CARMONA-ALDANA F, ZAMPEDRI C, SUASTE-OLMOS F, et al. CTCF knockout reveals an essential role for this protein during the zebrafish development [J]. Mechanisms of Development, 2018, 154: 51−59. DOI: 10.1016/j.mod.2018.04.006
[26] 肖成, 薛佳佳, 王晶, 等. 小尾寒羊S100A1基因编码区外显子克隆及表达分析 [J]. 中国畜牧杂志, 2021, 57(5):82−86. DOI: 10.19556/j.0258-7033.20200622-01 XIAO C, XUE J J, WANG J, et al. Cloning and expression analysis of exon of S100A1 gene in small tail Han sheep [J]. Chinese Journal of Animal Science, 2021, 57(5): 82−86.(in Chinese) DOI: 10.19556/j.0258-7033.20200622-01
[27] ZENG Z, HUANG N N, ZHANG Y D, et al. CTCF inhibits endoplasmic reticulum stress and apoptosis in cardiomyocytes by upregulating RYR2 via inhibiting S100A1 [J]. Life Sciences, 2020, 242: 117158. DOI: 10.1016/j.lfs.2019.117158
[28] LUO D, XIAO H W, DONG J L, et al. B7-H3 regulates lipid metabolism of lung cancer through SREBP1-mediated expression of FASN [J]. Biochemical and Biophysical Research Communications, 2017, 482(4): 1246−1251. DOI: 10.1016/j.bbrc.2016.12.021
[29] XU W W, CHEN Q M, JIA Y H, et al. Isolation, characterization, and SREBP1 functional analysis of mammary epithelial cell in buffalo [J]. Journal of Food Biochemistry, 2019, 43(11): e12997.
[30] BENGOECHEA-ALONSO M T, ERICSSON J. The phosphorylation-dependent regulation of nuclear SREBP1 during mitosis links lipid metabolism and cell growth [J]. Cell Cycle, 2016, 15(20): 2753−2765. DOI: 10.1080/15384101.2016.1220456
[31] MOISON C, CHAGRAOUI J, CARON M C, et al. Zinc finger protein E4F1 cooperates with PARP-1 and BRG1 to promote DNA double-strand break repair [J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(11): e2019408118. DOI: 10.1073/pnas.2019408118
[32] LIU Z Y, KRAUS W L. Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci [J]. Molecular Cell, 2017, 65(4): 589−603.e9. DOI: 10.1016/j.molcel.2017.01.017
[33] CERVANTES-CAMACHO C, BELTRÁN-LANGARICA A, OCHOA-URIBE A K, et al. The transient expression of Klf4 and Klf5 during adipogenesis depends on GSK3β activity [J]. Adipocyte, 2015, 4(4): 248−255. DOI: 10.1080/21623945.2015.1007823
[34] WU Q, FU C Y, LI M L, et al. CINP is a novel cofactor of KLF5 required for its role in the promotion of cell proliferation, survival and tumor growth [J]. International Journal of Cancer, 2019, 144(3): 582−594. DOI: 10.1002/ijc.31908
[35] WANG J, CHU Y F, XU M, et al. miR-21 promotes cell migration and invasion of hepatocellular carcinoma by targeting KLF5 [J]. Oncology Letters, 2019, 17(2): 2221−2227.
[36] SCHAAP F G, BINAS B, DANNEBERG H, et al. Impaired long-chain fatty acid utilization by cardiac myocytes isolated from mice lacking the heart-type fatty acid binding protein gene [J]. Circulation Research, 1999, 85(4): 329−337. DOI: 10.1161/01.RES.85.4.329
[37] YI B, WANG J G, WANG S, et al. Overexpression of Banna mini-pig inbred line fatty acid binding protein 3 promotes adipogenesis in 3T3-L1 preadipocytes [J]. Cell Biology International, 2014, 38(8): 918−923. DOI: 10.1002/cbin.10285
[38] CHO K H, KIM M J, JEON G J, et al. Association of genetic variants for FABP3 gene with back fat thickness and intramuscular fat content in pig [J]. Molecular Biology Reports, 2011, 38(3): 2161−2166. DOI: 10.1007/s11033-010-0344-3




 下载:
下载: