Genome-wide Identification and Expressions under Stresses of RLCK VI Family in Gossypium barbadense
-
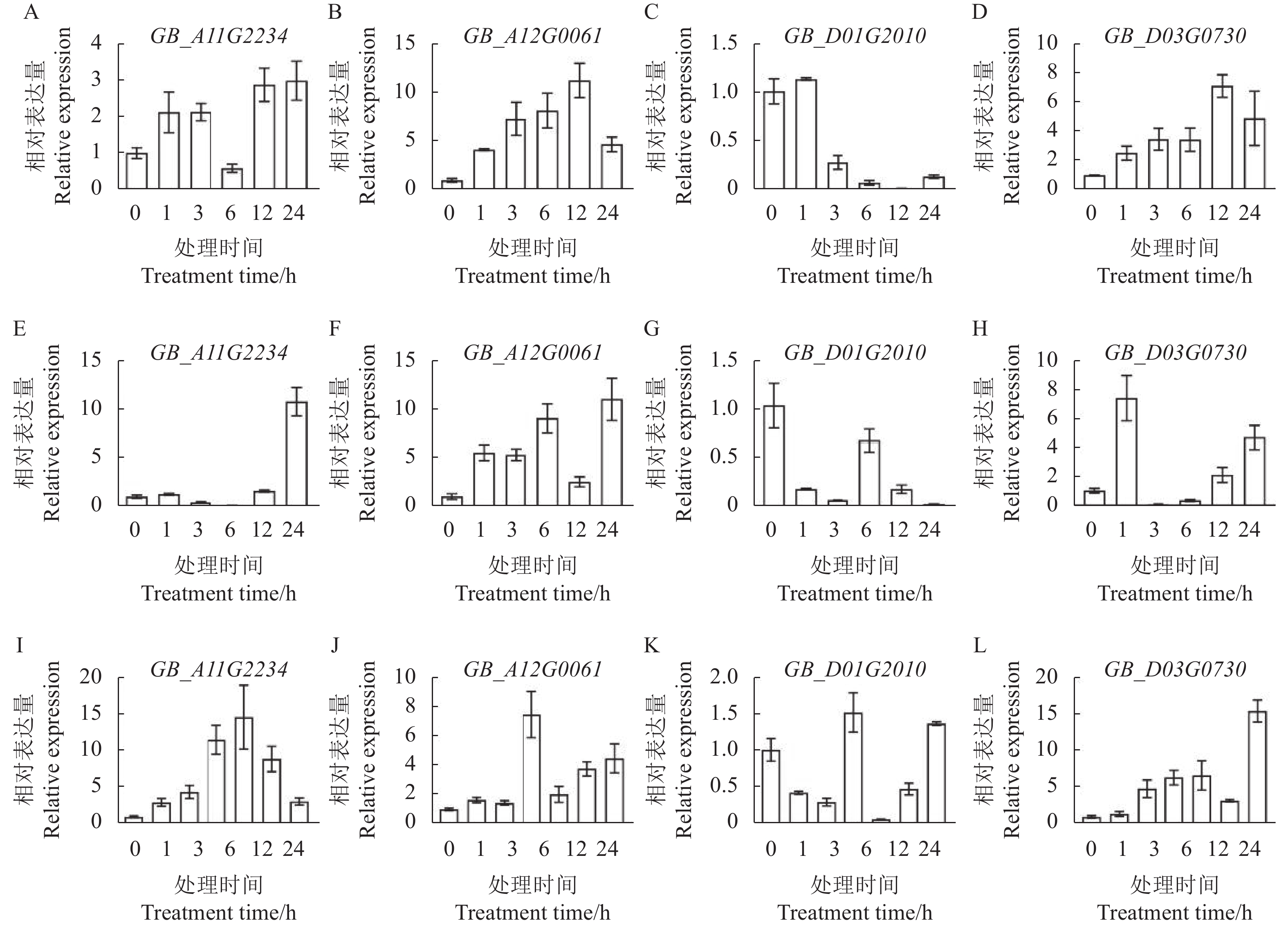
摘要:目的 对海岛棉(Gossypium barbadense)类受体胞质激酶RLCK VI(GbRLCK VI)家族基因进行全基因组分析,为深入研究 RLCK VI家族基因参与棉花生长发育和抗逆的调控机制提供参考。方法 基于最新发布的海岛棉基因组数据,利用生物信息学手段对GbRLCK VI家族基因进行全基因组鉴定,并系统分析该家族基因成员的理化性质、序列特征、基因复制、系统进化和表达特征。结果 海岛棉中共鉴定出39个GbRLCK VI家族基因,经聚类分析将其分为A、B两组,其中A组22个,B组17个,均含有1个激酶结构域,分布于16条染色体,多数位于质膜。基因复制分析表明,该家族在进化过程中发生染色体片段复制事件;Ka/Ks分析显示,所有基因对Ka/Ks均小于1,表明GbRLCK VI家族基因在进化过程中可能经历了严格的纯化选择作用。转录组分析表明,GbRLCK VI家族基因在10个不同组织中的表达模式不同,11个基因在花器官中优势表达,9个基因在根茎叶中优势表达;逆境胁迫下的表达分析显示,8个基因在干旱、盐、黄萎病胁迫下优势表达,4个基因只在黄萎病胁迫下优势表达,说明GbRLCK VI基因能快速地参与抗逆反应,挑选4个基因GB_A12G0061、GB_A11G2234、GB_D01G2010、GB_D03G0730进行qRT-PCR验证,表达分析结果显示,4个基因在干旱、盐或黄萎病菌胁迫下的表达趋势与转录组数据一致,表明它们参与了棉花对逆境胁迫(干旱、盐和黄萎病菌)的响应过程。结论 明确了GbRLCK VI家族基因在基因组中的分布特征、结构特征以及系统进化特征,根据转录组数据初步揭示了该家族基因在棉花生长发育和抗逆胁迫中的功能。Abstract:Objective Genomes and expressions under stresses of the RLCK VI family genes in Gossypium barbadense were determined to study the regulatory mechanisms of the growth, development, and stress resistance of cotton plants.Method Based on the latest released data on G. barbadense genome, bioinformatics of GbRLCK VI was analyzed to understand the associated physiochemical properties, sequence characteristics, gene replication, phylogenetic evolution, and expression.Result Thirty-nine RLCK VI were identified in G. barbadense which were clustered into two categories of 22 in Group A and 17 in Group B. Both groups contained a kinase domain distributed in 16 chromosomes with most of them located in plasma membrane. The gene family had undergone chromosome fragment duplication events during evolution. Since all Ka/Ks of the gene pairs were less than 1, strict purification and selection might have taken place in the process. The expressions of GbRLCK VI as shown by the transcriptome analysis varied in 10 different tissues with 11 predominantly expressed in the floral organs, while 9 in the roots, stems, and leaves. Under different stresses, 8 genes were significantly expressed by the imposed drought, salt, and verticillium wilt, and 4 only by verticillium wilt. On 4 selected genes, i.e., GB_ A12G0061, GB_ A11G2234, GB_ D01G2010, and GB_ D03G0730, qRT-PCR showed their expressions under drought, salt, or verticillium wilt stress to agree with what the transcriptome data did. Their involvement in the stress response of the cotton plant was confirmed.Conclusion The genome, structure, and phylogenetic characteristics of GbRLCK VI family in G. barbadense were determined. Their roles in the growth, development, and stress responses of the cotton plant were clarified.
-
烟粉虱Bemisia tabaci是世界性的重要害虫,寄主广泛,可通过直接刺吸、分泌蜜露诱发煤烟病以及传播病毒等产生危害[1]。由于烟粉虱的防治过分依赖化学杀虫剂,导致烟粉虱对大部分杀虫剂产生了抗药性,增加了防治难度[2-5]。因此,有必要寻求一些相应的替代措施,以缓解烟粉虱抗药性的发展。
长期以来,烟粉虱的生物防治研究一直备受关注,利用虫生真菌、捕食性天敌、寄生性天敌的生物防治成为替代化学防治的最重要手段之一[6-11]。我国烟粉虱的天敌资源丰富,其中日本刀角瓢虫Serangium japonicum是烟粉虱的一种重要的本地捕食性天敌优势种,广泛分布在我国南方地区[12-13]。但关于日本刀角瓢虫对烟粉虱控制效应的研究仍较少见[14-15]。本研究在实验室条件和田间笼罩实验条件下,测定了日本刀角瓢虫对烟粉虱的捕食能力及其不同释放密度对烟粉虱种群的控制效果,以期了解日本刀角瓢虫田间释放对烟粉虱种群的控制潜能,为有效利用该天敌瓢虫进行烟粉虱的生物防治提供依据。
1. 材料与方法
1.1 供试昆虫
烟粉虱和日本刀角瓢虫均是田间采集并室内饲养的实验种群。
1.2 日本刀角瓢虫对烟粉虱的捕食量测定
日本刀角瓢虫雌成虫对烟粉虱不同虫态的捕食量:在室内盆栽的茄子叶上定量接入烟粉虱成虫,让其产卵24 h,然后剔除成虫,饲养不同虫态的烟粉虱。按烟粉虱不同的发育进度取带有卵、1~4龄若虫的叶片在解剖镜下计数后在培养皿中饲喂单头瓢虫,每皿投入饥饿24 h的瓢虫成虫1头,24 h后调查记录被捕食猎物数量,每个处理重复10次。
日本刀角瓢虫各虫态对烟粉虱卵的捕食量:测定在直径为110 mm的培养皿中进行,每皿烟粉虱卵800~1 000粒。每皿分别饲喂瓢虫单头雌雄成虫、1~4龄幼虫,24 h后检查各处理被捕食卵粒数,每个处理重复10次。
1.3 日本刀角瓢虫成虫对烟粉虱卵的功能反应测定
在直径为110 mm的培养皿中进行测定,烟粉虱卵密度设为每皿100、200、400、600、800、1 000、1 200粒卵。每皿投入饥饿24 h的刀角瓢虫雌成虫1头,后置于温度27℃、相对湿度65%和光照14:10(L:D)的培养箱内,24 h后检查各处理被捕食卵粒数,重复10次。统计捕食量并拟合功能反应曲线。
1.4 日本刀角瓢虫成虫对烟粉虱种群的控制效果
试验于盆栽茄子苗在田间笼罩(80 cm×80 cm×150 cm)条件下进行,每盆1株,每笼罩放3盆。每株茄子苗上接入烟粉虱成虫150对,接虫4周后释放瓢虫,释放密度设置4个处理,分别为不释放瓢虫(对照处理,CK)、每株1、3、5对瓢虫,每个处理重复3次。每周调查1次不同处理中烟粉虱的种群数量消长动态,每盆苗每次调查其中1株的第1片叶、第2株苗的第4片叶和第3株的第7片叶,每周轮换抽叶调查,剪叶前先调查每片叶背面烟粉虱成虫数量,然后在体视镜下检查每片剪叶上1 cm2卵+若虫(伪蛹视为四龄若虫)的数量,每片叶上随机检查3个1 cm2,并将所调查的同株3片叶片上烟粉虱成虫或卵+若虫的平均值作为该处理的观察值。共调查10周,并根据第10周的调查结果计算瓢虫不同释放密度对烟粉虱种群的控制效果。
控制效果/%=[(对照区种群数量-释放区种群数量) /对照区种群数量]×100%
1.5 数据处理
捕食功能反应采用HollingⅡ型圆盘方程[16]进行拟合,其方程式为:
Ne=α·T·N0/(1+α·Th·N0)
其中:Ne为被捕食的猎物数;N0为猎物密度;T为试验总用时(在本研究中T=1 d);α为瞬间攻击率;Th为处置时间。根据所拟合的捕食功能方程参数α和Th,计算瓢虫对烟粉虱卵的日最大捕食量。采用单因素方差分析的方法对捕食量数据进行分析,采用重复测量的方法对瓢虫对烟粉虱种群的控制效应进行分析,数据分析在R语言[17]软件上进行。
2. 结果与分析
2.1 日本刀角瓢虫对烟粉虱的捕食量
图 1显示,日本刀角瓢虫雌成虫对烟粉虱的卵和各龄若虫均表现出很强的捕食能力,其中对烟粉虱卵的捕食量最高,单雌日均捕食量可达640.8粒;对烟粉虱各龄若虫的捕食量由高到低依次为1龄若虫、2龄若虫、3龄若虫、4龄若虫日均捕食量分别为514.4、306.0、136.1、70.4头,瓢虫对烟粉虱卵和各龄若虫的捕食量之间差异显著(F4,45 =127.6,P < 0.001)。图 2显示,不同虫态日本刀角瓢虫对烟粉虱卵的捕食量差异显著(F5,54=130.8,P < 0.001),瓢虫雌雄成虫及高龄幼虫对烟粉虱卵具有较强的捕食能力,其中4龄幼虫的捕食量最高,单头日均捕食量可达660.9粒,1龄幼虫的捕食量最低,日均捕食量为57.8粒。
![]() 图 4 释放日本刀角瓢虫对烟粉虱成虫数量消长动态的影响注:T1为释放1对瓢虫,T3为释放3对瓢虫,T5为释放5对瓢虫;箭头指示处为瓢虫释放时间,即调查的第4周。图 5同。Figure 4. Effect of S. japonicum release on population of adult B. tabaci
图 4 释放日本刀角瓢虫对烟粉虱成虫数量消长动态的影响注:T1为释放1对瓢虫,T3为释放3对瓢虫,T5为释放5对瓢虫;箭头指示处为瓢虫释放时间,即调查的第4周。图 5同。Figure 4. Effect of S. japonicum release on population of adult B. tabaci2.2 日本刀角瓢虫成虫对烟粉虱卵的功能反应
图 3显示,日本刀角瓢虫雌成虫对烟粉虱卵的功能反应模型属HollingⅡ型。利用HollingⅡ型圆盘方程拟合试验数据,求得日本刀角瓢虫雌成虫对烟粉虱卵的功能反应参数,结果显示,日本刀角瓢虫雌成虫的瞬间攻击率(α)为1.014 4(95%置信限为0.964 4~1.064 4),处理猎物时间(Th)为0.000 7(95%置信限为0.000 6~0.000 9),最大日捕食量(1/h)为1366.78粒。
2.3 日本刀角瓢虫对烟粉虱种群的控制作用
图 4结果显示,释放瓢虫前4周,即总调查时间的第1~4周,各处理间烟粉虱成虫数量差异不显著(F3,8 =1.131,P=0.393);释放瓢虫3周后,各处理间烟粉虱成虫数量差异也不显著(F3,8 =2.104,P=0.178);每株释放3对和5对瓢虫处理的烟粉虱成虫数量在释放瓢虫后的第4~6周均显著低于对照处理,而每株释放1对瓢虫处理的烟粉虱成虫数量在释放瓢虫后第4周与对照处理差异不显著,释放后第5周才出现明显下降。释放瓢虫6周后,不同处理间烟粉虱成虫的数量差异显著(F3,8 =40.601,P < 0.001),释放瓢虫处理的烟粉虱成虫数量均显著少于对照处理(T1:CK:z=3.807,P < 0.001;T3:CK:z=3.976,P < 0.001;T5:CK:z=5.715,P < 0.001)。释放瓢虫后的第6周,每株释放1、3、5对瓢虫处理的烟粉虱成虫数量分别为30.44、18.22、10.67头·cm-2,均显著低于对照处理的烟粉虱成虫数量(68.11头·cm-2),控制效果分别达55.31%、73.25%和84.33%。
图 5显示,释放瓢虫前4周,即总调查时间的第1~4周,各处理间烟粉虱卵+若虫的数量差异不显著(F3,8 =0.976,P=0.451);释放瓢虫1周后,各处理间烟粉虱卵+若虫的数量差异也不显著(F3,8=2.335,P=0.150);释放瓢虫6周后,各处理间烟粉虱卵+若虫的数量差异显著(F3,8=549.693,P < 0.001),释放瓢虫处理的烟粉虱卵+若虫的数量显著少于对照处理(T1:CK:z=3.799,P < 0.001;T3:CK:z=9.704,P < 0.001;T5:CK:z=11.215,P < 0.001),释放3对瓢虫(z=3.448,P=0.003)和释放5对瓢虫(z=4.059,P < 0.001)处理的控制效果显著优于释放1对瓢虫处理,而释放3对瓢虫和释放5对瓢虫的处理间差异不显著(z=0.688,P=0.897)。每株释放3对和5对瓢虫处理的烟粉虱卵+若虫数量在释放瓢虫后的第2周开始下降,而每株释放1对瓢虫处理的烟粉虱卵+若虫数量在释放瓢虫后的第2周仍持续增长,并在释放瓢虫后的第4周达到高峰,此后开始下降,瓢虫的控制效果才逐步显现。在释放瓢虫后的第6周,每株释放1、3、5对瓢虫处理的烟粉虱卵+若虫数量分别为36.22、13.22、6.44头·cm-2,均显著低于对照处理的烟粉虱非成虫数量(78.43头·cm-2),控制效果分别达53.82%、83.14%和91.79%。
3. 讨论与结论
捕食量是测定捕食性天敌的捕食潜能的一个重要指标。本研究结果发现,日本刀角瓢虫各龄幼虫和成虫均可取食烟粉虱卵和若虫,而且表现出很强的捕食能力,尤其对烟粉虱卵的捕食量巨大。日本刀角瓢虫4龄幼虫和雌成虫对烟粉虱卵的日均捕食量分别高达660.9粒·头-1和640.8粒·雌-1,这与姚松林等[14]的研究结果基本一致,因此日本刀角瓢虫对烟粉虱具有较好的控制潜力,在生物防治上极具开发应用价值。
田间笼罩试验结果发现,在瓢虫释放后的初期,烟粉虱成虫和卵+若虫数量没有被有效控制,而是与对照处理一样在此后的1~4周仍然继续增长,其中每株释放1对瓢虫处理的成虫和卵+若虫数量均在释放后第4周达到高峰,随后才逐步表现出对烟粉虱种群良好的控制效果;每株释放3对和5对瓢虫处理的成虫数量在释放后第3周达到高峰,卵+若虫数量在释放后第1~2周达到高峰,随后逐步表现出对烟粉虱种群的控制效果,表明释放日本刀角瓢虫对烟粉虱种群数量控制存在滞后现象。究其原因主要是:在释放瓢虫后的初期,由于瓢虫可直接捕食烟粉虱卵和若虫,而且捕食能力很强,使其对烟粉虱卵+若虫数量的控制效应可以在较短的时间内体现出来。但是,前期接入的150对烟粉虱经过4周时间的繁殖已经建立了较高的种群密度,在瓢虫释放前的抽样调查中发现分布于茄子植株中下部叶片上的大量烟粉虱拟蛹正在陆续羽化,导致烟粉虱成虫种群数量在瓢虫释放后的前几周内仍然可以继续增长。释放后期对烟粉虱成虫数量的控制作用则与瓢虫定居后的持续捕食、成功繁殖和种群增长有密切关系,因为在释放瓢虫后的第2周开始陆续观察到瓢虫新孵育的卵和幼虫,而且随后几周瓢虫种群数量不断增加。在释放瓢虫后的第4周开始,烟粉虱成虫的数量受到明显抑制。10周时,释放瓢虫显著地控制了烟粉虱种群密度,每株释放3对和5对瓢虫对烟粉虱成虫种群的控制效果分别高达73.25%和84.33%。因此,在生物防治中天敌释放后能否成功定居、繁殖并建立种群,对释放天敌对目标害虫的控制效果和持续控制是至关重要的。
已有研究表明,日本刀角瓢虫成虫寿命长,在26℃下产卵前期为3~10 d,不同羽化日龄成虫所处的生殖状态差异明显(另文发表)。在本研究的田间笼罩试验中,所释放瓢虫成虫的生殖状态至关重要,它将直接影响瓢虫释放后的种群建立及对烟粉虱种群的持续控制效果。此外,烟粉虱的生长发育和种群繁殖与湿度条件密切相关[18],本研究过程中笼罩内较高的湿度条件也会对试验结果产生一定的影响。因此,日本刀角瓢虫对烟粉虱自然种群的控制效果有待进一步的验证。
-
图 1 拟南芥、水稻与海岛棉RLCK VI家族基因系统进行分析
红色方块为RLCK VI_A亚家族;绿色方块为RLCK VI_B亚家族;At:拟南芥,LOC_Os:水稻,GB:海岛棉。
Figure 1. Phylogenetic evolution of RLCK VI family genes in Arabidopsis thaliana, Oryza sativa, and G. barbadense
Red square indicates RLCK VI_A subfamily group; green square, RLCK VI_B subfamily group; At: Arabidopsis thaliana; LOC_Os: Oryza sativa; GB: G. barbadense.
表 1 荧光定量 PCR引物序列
Table 1 Sequences of primers for quantitative PCR
基因名称
Gene name基因序列(5′-3′)
Primer sequence (5′-3′)GB_A11G2234-F AATGAAGAATGAGAAACAA GB_A11G2234-R GAGGTGAAAACTGAAGTAC GB_A12G0061-F AAACTGGACTCACCACAAC GB_A12G0061-R AGTACACCAAAGGCAAACA GB_D01G2010-F GCAATATGGGGACCAACTG GB_D01G2010-R AGAACAACACCGAAAGCGT GB_D03G0730-F CATAAACGAAATAGCTTGC GB_D03G0730-R CCTTGGTCTCATAGGAAAC GhUBQ7-F GAAGGCATTCCACCTGACCAAC GhUBQ7-R CTTGACCTTCTTCTTCTTGTGCTTG 表 2 GbRLCK VI亚族成员蛋白理化性质及亚细胞定位分析
Table 2 Physicochemical properties and subcellular localizations of proteins in GbRLCK VI subfamily members
基因编号
Gene ID分类
Classification理论等电点
pI相对分子质量
Relative molecular
mass/Da染色体
Chromosome位置
Location/bp开放阅读
框长度
ORF/bp氨基酸
Amino
acids/aa亚细胞定位
Subcellular
localizationGB_A01G0260 RLCK VI_A 5.30 62875.09 A 2206175~2210244 1683 561 细胞质 Cytoplasmic GB_A01G1081 RLCK VI_B 8.18 83349.28 A 20600106~20603458 2214 738 胞外 Extracellular GB_A01G1914 RLCK VI_B 5.97 75989.42 A 103561645~103564486 2046 682 胞外 Extracellular GB_A02G1246 RLCK VI_B 6.94 84611.20 A 63519857~63524024 2325 775 胞外 Extracellular GB_A02G1740 RLCK VI_A 5.94 50565.48 A 97801526~97803417 1362 454 质膜 Plasma membrane GB_A03G0311 RLCK VI_A 5.95 44313.24 A 3766673~3768994 1167 389 质膜 Plasma membrane GB_A05G3702 RLCK VI_A 8.52 67781.48 A 84140713~84143610 1815 605 细胞质 Cytoplasmic GB_A08G1010 RLCK VI_A 9.08 65230.47 A 33774846~33777243 1749 583 细胞质 Cytoplasmic GB_A09G2643 RLCK VI_A 5.6 58175.80 A 77782790~77786063 1554 518 细胞质 Cytoplasmic GB_A10G1115 RLCK VI_A 6.01 54570.32 A 22829603~22832184 1470 490 质膜 Plasma membrane GB_A11G0540 RLCK VI_A 6.03 50910.97 A 4885422~4887763 1362 454 质膜 Plasma membrane GB_A11G1251 RLCK VI_A 9.25 47356.59 A 12738544~12740333 1269 423 质膜 Plasma membrane GB_A11G2234 RLCK VI_A 6.09 52234.38 A 51930366~51932808 1395 465 质膜 Plasma membrane GB_A11G2998 RLCK VI_A 6.98 54619.72 A 102734569~102739375 1455 485 质膜 Plasma membrane GB_A11G3462 RLCK VI_A 8.39 54509.86 A 111382556~111386884 1455 485 细胞质 Cytoplasmic GB_A12G0061 RLCK VI_A 9.54 44738.44 A 770437~772212 1197 399 质膜 Plasma membrane GB_A12G0366 RLCK VI_B 5.53 51025.69 A 6110842~6112604 1365 455 质膜 Plasma membrane GB_A12G0580 RLCK VI_B 8.94 57833.28 A 12489405~12491904 1566 522 胞外 Extracellular GB_A12G0837 RLCK VI_B 6.28 72794.02 A 35264457~35267552 1959 653 胞外 Extracellular GB_A12G1016 RLCK VI_A 6.08 55330.70 A 58620348~58623557 1470 490 细胞质 Cytoplasmic GB_D01G0255 RLCK VI_A 5.61 54753.77 D 2136137~2138668 1461 487 质膜 Plasma membrane GB_D01G1159 RLCK VI_B 8.62 82638.79 D 16772231~16775594 2193 731 胞外 Extracellular GB_D01G2010 RLCK VI_B 5.87 75966.19 D 54299320~54302161 2049 683 胞外 Extracellular GB_D03G0345 RLCK VI_A 5.94 50468.36 D 3912795~3914684 1359 453 质膜 Plasma membrane GB_D03G0730 RLCK VI_B 6.52 84629.22 D 19203868~19208089 2328 776 胞外 Extracellular GB_D03G1685 RLCK VI_A 5.86 44338.23 D 49960463~49962648 1167 389 质膜 Plasma membrane GB_D04G0916 RLCK VI_A 8.82 67857.57 D 17923478~17926392 1824 608 细胞质 Cytoplasmic GB_D09G2477 RLCK VI_A 5.54 58118.73 D 52528068~52531290 1554 518 细胞质 Cytoplasmic GB_D10G1818 RLCK VI_A 6.1 58069.37 D 46511912~46514475 1569 523 细胞质 Cytoplasmic GB_D11G0554 RLCK VI_A 5.96 51742.65 D 4472367~4474758 1386 462 质膜 Plasma membrane GB_D11G1285 RLCK VI_A 9.37 50810.57 D 11385965~11387753 1347 449 细胞质 Cytoplasmic GB_D11G2293 RLCK VI_A 6.17 52301.37 D 30637528~30639969 1395 465 质膜 Plasma membrane GB_D11G2991 RLCK VI_A 7.67 54466.53 D 60438857~60443672 1455 485 质膜 Plasma membrane GB_D11G3433 RLCK VI_A 8.39 54454.82 D 67010248~67014591 1455 485 细胞质 Cytoplasmic GB_D12G0064 RLCK VI_A 9.51 44758.54 D 777708~781902 1197 399 质膜 Plasma membrane GB_D12G0350 RLCK VI_B 5.54 51097.86 D 4562988~4564741 1365 455 质膜 Plasma membrane GB_D12G0573 RLCK VI_B 9.04 57727.16 D 9309237~9311733 1563 521 胞外 Extracellular GB_D12G0968 RLCK VI_A 5.73 53551.68 D 20078236~20080755 1425 475 细胞质 Cytoplasmic GB_D12G0998 RLCK VI_B 6.62 79099.37 D 16414879~16417975 2133 711 胞外 Extracellular 表 3 串联重复基因Ka/Ks计算
Table 3 Calculation of Ka/Ks for tandem repeat gene
基因编号
Gene ID基因编号
Gene ID非同义替换
Ka同义替换
Ks非同义替换/同义替换
Ka/KsGB_A01G1914 GB_A12G0366 0.104312114 0.45263137 0.230457101 GB_A01G0260 GB_D01G0255 0.028943225 0.046322927 0.624814254 GB_A01G1081 GB_D01G1159 0.020977245 0.054445666 0.385287692 GB_A01G1914 GB_D01G2010 0.013406752 0.02845499 0.471156428 GB_A01G1914 GB_D12G0350 0.109277637 0.473782321 0.230649461 GB_A02G1740 GB_D03G0345 0.005691277 0.03053857 0.186363574 GB_A02G1246 GB_D03G0730 0.006776517 0.037543027 0.18050001 GB_A02G1740 GB_D11G2293 0.197554085 1.040010514 0.18995393 GB_A05G3702 GB_D04G0916 0.023308504 0.056770511 0.410574135 GB_A09G2643 GB_D09G2477 0.012551719 0.050376868 0.249156396 GB_A10G1115 GB_D09G2477 0.250706504 0.744111803 0.336920478 GB_A10G1115 GB_D10G1818 0.010688224 0.02700967 0.395718402 GB_A11G1251 GB_A12G0061 0.103086568 0.88836622 0.116040621 GB_A11G0540 GB_D11G0554 0.01383297 0.035350161 0.391312788 GB_A11G1251 GB_D11G1285 0.012356552 0.038946803 0.31726743 GB_A11G2998 GB_D11G2991 0.01397591 0.054049622 0.258575533 GB_A11G3462 GB_D11G3433 0.013455287 0.069780882 0.192821966 GB_A11G0540 GB_D11G2293 0.239191447 0.679802163 0.351854496 GB_A11G2234 GB_D11G2293 0.009270106 0.04321328 0.214519851 GB_A11G1251 GB_D12G0064 0.108697399 0.818974304 0.132723821 GB_A12G0366 GB_D01G2010 0.110134899 0.497372712 0.221433336 GB_A12G0061 GB_D11G1285 0.102473606 0.89384392 0.114643735 GB_A12G0061 GB_D12G0064 0.017583223 0.05245583 0.335200547 GB_A12G0580 GB_D12G0573 0.006683419 0.045728447 0.146154523 GB_A12G0366 GB_D12G0350 0.01232937 0.026976777 0.457036417 GB_A12G1016 GB_D12G0968 0.014602332 0.065561783 0.222726282 GB_D01G2010 GB_D12G0350 0.116240086 0.513096743 0.22654614 GB_D03G0345 GB_D11G2293 0.19515733 1.062230892 0.18372402 GB_D09G2477 GB_D10G1818 0.254053518 0.74467519 0.341160175 GB_D11G0554 GB_D11G2293 0.210367409 0.660727812 0.318387398 GB_D11G1285 GB_D12G0064 0.106823583 0.823959217 0.129646687 -
[1] 易黎. 拟南芥及甘蓝型油菜RBK2蛋白及相关蛋白家族生物信息学分析[D]. 郑州: 郑州大学, 2016. YI L. Bioinformatics analysis of RBK2 and its related protein family in Arabidopsis thaliana and Braasica napus[D]. Zhengzhou: Zhengzhou University, 2016. (in Chinese)
[2] 饶绍飞. 拟南芥类受体胞质激酶第七亚家族成员在先天免疫中的功能分析[D]. 北京: 中国科学院大学, 2018. RAO S F. Functional analysis of members of the seventh subfamily of Arabidopsis receptor cytoplasmic kinases in innate immunity[D]. Beijing: University of Chinese Academy of Sciences, 2018. (inChinese)
[3] VIJ S, GIRI J, DANSANA P K, et al. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: Organization, phylogenetic relationship, and expression during development and stress [J]. Molecular Plant, 2008, 1(5): 732−750. DOI: 10.1093/mp/ssn047
[4] REINER T, HOEFLE C, HUESMANN C, et al. The Arabidopsis ROP-activated receptor-like cytoplasmic kinase RLCK VI_A3 is involved in control of basal resistance to powdery mildew and trichome branching [J]. Plant Cell Reports, 2015, 34(3): 457−468. DOI: 10.1007/s00299-014-1725-1
[5] 马银花, 李萍芳, 董文静, 等. 水稻抗性蛋白OsRRK1抗褐飞虱机理分析 [J]. 中国水稻科学, 2020, 34(6):512−519. DOI: 10.16819/j.1001-7216.2020.0406 MA Y H, LI P F, DONG W J, et al. Mechanism analysis of rice resistance protein OsRRK1 against the brown planthopper [J]. Chinese Journal of Rice Science, 2020, 34(6): 512−519.(in Chinese) DOI: 10.16819/j.1001-7216.2020.0406
[6] 何含杰, 张党权, 唐丽, 等. 植物RLCK的生物学功能与信号途径研究进展 [J]. 植物生理学报, 2014, 50(7):885−890. DOI: 10.13592/j.cnki.ppj.2014.0154 HE H J, ZHANG D Q, TANG L, et al. Recent advance on biological function and signal pathway of receptor-like cytoplasmic kinase in plants [J]. Plant Physiology Journal, 2014, 50(7): 885−890.(in Chinese) DOI: 10.13592/j.cnki.ppj.2014.0154
[7] COSTA A T, BRAVO J P, KRAUSE-SAKATE R, et al. The receptor-like kinase SlSOBIR1 is differentially modulated by virus infection but its overexpression in tobacco has no significant impact on virus accumulation [J]. Plant Cell Reports, 2016, 35(1): 65−75. DOI: 10.1007/s00299-015-1868-8
[8] JURCA M E, BOTTKA S, FEHÉR A. Characterization of a family of Arabidopsis receptor-like cytoplasmic kinases (RLCK class VI) [J]. Plant Cell Reports, 2008, 27(4): 739−748. DOI: 10.1007/s00299-007-0494-5
[9] JUNG K H, CAO P J, SEO Y S, et al. The Rice Kinase Phylogenomics Database: A guide for systematic analysis of the rice kinase super-family [J]. Trends in Plant Science, 2010, 15(11): 595−599. DOI: 10.1016/j.tplants.2010.08.004
[10] LEE L Y C, HOU X L, FANG L, et al. STUNTED mediates the control of cell proliferation by GA in Arabidopsis [J]. Development, 2012, 139(9): 1568−1576. DOI: 10.1242/dev.079426
[11] VALKAI I, KÉNESI E, DOMONKOS I, et al. The Arabidopsis RLCK VI_A2 kinase controls seedling and plant growth in parallel with gibberellin [J]. International Journal of Molecular Sciences, 2020, 21(19): 7266. DOI: 10.3390/ijms21197266
[12] ENDERS T A, FRICK E M, STRADER L C. An Arabidopsis kinase cascade influences auxin-responsive cell expansion [J]. The Plant Journal, 2017, 92(1): 68−81. DOI: 10.1111/tpj.13635
[13] LAL N K, FISHER A J, DINESH-KUMAR S P. Arabidopsis receptor-like cytoplasmic kinase BIK1: Purification, crystallization and X-ray diffraction analysis[J]. Acta Crystallographica Section F, Structural Biology Communications, 2016, 72(Pt 10): 738-742.
[14] LU D P, WU S J, GAO X Q, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity [J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(1): 496−501. DOI: 10.1073/pnas.0909705107
[15] HUESMANN C, REINER T, HOEFLE C, et al. Barley ROP binding kinase1 is involved in microtubule organization and in basal penetration resistance to the barley powdery mildew fungus [J]. Plant Physiology, 2012, 159(1): 311−320. DOI: 10.1104/pp.111.191940
[16] 马银花, 莫凯琴, 刘璐, 等. 过量表达OsRRK1对水稻叶片发育的影响 [J]. 中国农业科学, 2021, 54(5):877−886. DOI: 10.3864/j.issn.0578-1752.2021.05.001 MA Y H, MO K Q, LIU L, et al. Effect of overexpression of OsRRK1 gene on rice leaf development [J]. Scientia Agricultura Sinica, 2021, 54(5): 877−886.(in Chinese) DOI: 10.3864/j.issn.0578-1752.2021.05.001
[17] 田超, 王冉, 彭艳, 等. 植物抗逆胁迫相关蛋白激酶的研究进展 [J]. 安徽农业科学, 2015, 43(20):4−6,37. DOI: 10.3969/j.issn.0517-6611.2015.20.002 TIAN C, WANG R, PENG Y, et al. Research advance of protein kinase in plant resistant to adversity stress [J]. Journal of Anhui Agricultural Sciences, 2015, 43(20): 4−6,37.(in Chinese) DOI: 10.3969/j.issn.0517-6611.2015.20.002
[18] 赵曾强, 孙国清, 张国丽, 等. 海岛棉GbRLCK10基因克隆及表达分析 [J]. 西北植物学报, 2017, 37(11):2130−2138. DOI: 10.7606/j.issn.1000-4025.2017.11.2130 ZHAO Z Q, SUN G Q, ZHANG G L, et al. Cloning and expression analysis of the GbRLCK10 gene in Gossypium barbadense L. [J]. Acta Botanica Boreali-Occidentalia Sinica, 2017, 37(11): 2130−2138.(in Chinese) DOI: 10.7606/j.issn.1000-4025.2017.11.2130
[19] RAMEGOWDA V, BASU S, KRISHNAN A, et al. Rice growth under drought kinase is required for drought tolerance and grain yield under normal and drought stress conditions [J]. Plant Physiology, 2014, 166(3): 1634−1645. DOI: 10.1104/pp.114.248203
[20] SUN X L, SUN M Z, LUO X, et al. A Glycine soja ABA-responsive receptor-like cytoplasmic kinase, GsRLCK, positively controls plant tolerance to salt and drought stresses [J]. Planta, 2013, 237(6): 1527−1545. DOI: 10.1007/s00425-013-1864-6
[21] DORJGOTOV D, JURCA M E, FODOR-DUNAI C, et al. Plant Rho-type (Rop) GTPase-dependent activation of receptor-like cytoplasmic kinases in vitro [J]. FEBS Letters, 2009, 583(7): 1175−1182. DOI: 10.1016/j.febslet.2009.02.047
[22] AGRAWAL G K, IWAHASHI H, RAKWAL R. Small GTPase ‘Rop’: Molecular switch for plant defense responses [J]. FEBS Letters, 2003, 546(2/3): 173−180.
[23] HU Y, CHEN J D, FANG L, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton [J]. Nature Genetics, 2019, 51(4): 739−748. DOI: 10.1038/s41588-019-0371-5
[24] CHEN C J, CHEN H, ZHANG Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data [J]. Molecular Plant, 2020, 13(8): 1194−1202. DOI: 10.1016/j.molp.2020.06.009
[25] KUMAR S, STECHER G, TAMURA K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets [J]. Molecular Biology and Evolution, 2016, 33(7): 1870−1874. DOI: 10.1093/molbev/msw054
[26] VERA ALVAREZ R, PONGOR L S, MARIÑO-RAMÍREZ L, et al. TPMCalculator: One-step software to quantify mRNA abundance of genomic features [J]. Bioinformatics, 2019, 35(11): 1960−1962. DOI: 10.1093/bioinformatics/bty896
[27] SHABAN M, MIAO Y H, ULLAH A, et al. Physiological and molecular mechanism of defense in cotton against Verticillium dahliae [J]. Plant Physiology and Biochemistry, 2018, 125: 193−204. DOI: 10.1016/j.plaphy.2018.02.011
[28] WANG M J, TU L L, YUAN D J, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense [J]. Nature Genetics, 2019, 51(2): 224−229. DOI: 10.1038/s41588-018-0282-x
[29] 庞丹丹, 刘玉飞, 田易萍, 等. 茶树ZF-HD转录因子基因家族的鉴定及表达分析 [J]. 南方农业学报, 2021, 52(3):632−640. DOI: 10.3969/j.issn.2095-1191.2021.03.011 PANG D D, LIU Y F, TIAN Y P, et al. Identification and expression analysis of ZF-HD transcription factor gene family in Camellia sinensis [J]. Journal of Southern Agriculture, 2021, 52(3): 632−640.(in Chinese) DOI: 10.3969/j.issn.2095-1191.2021.03.011





 下载:
下载: