Mitochondrial Genome and Phylogeny of Bactrocera (B.) tuberculata
-
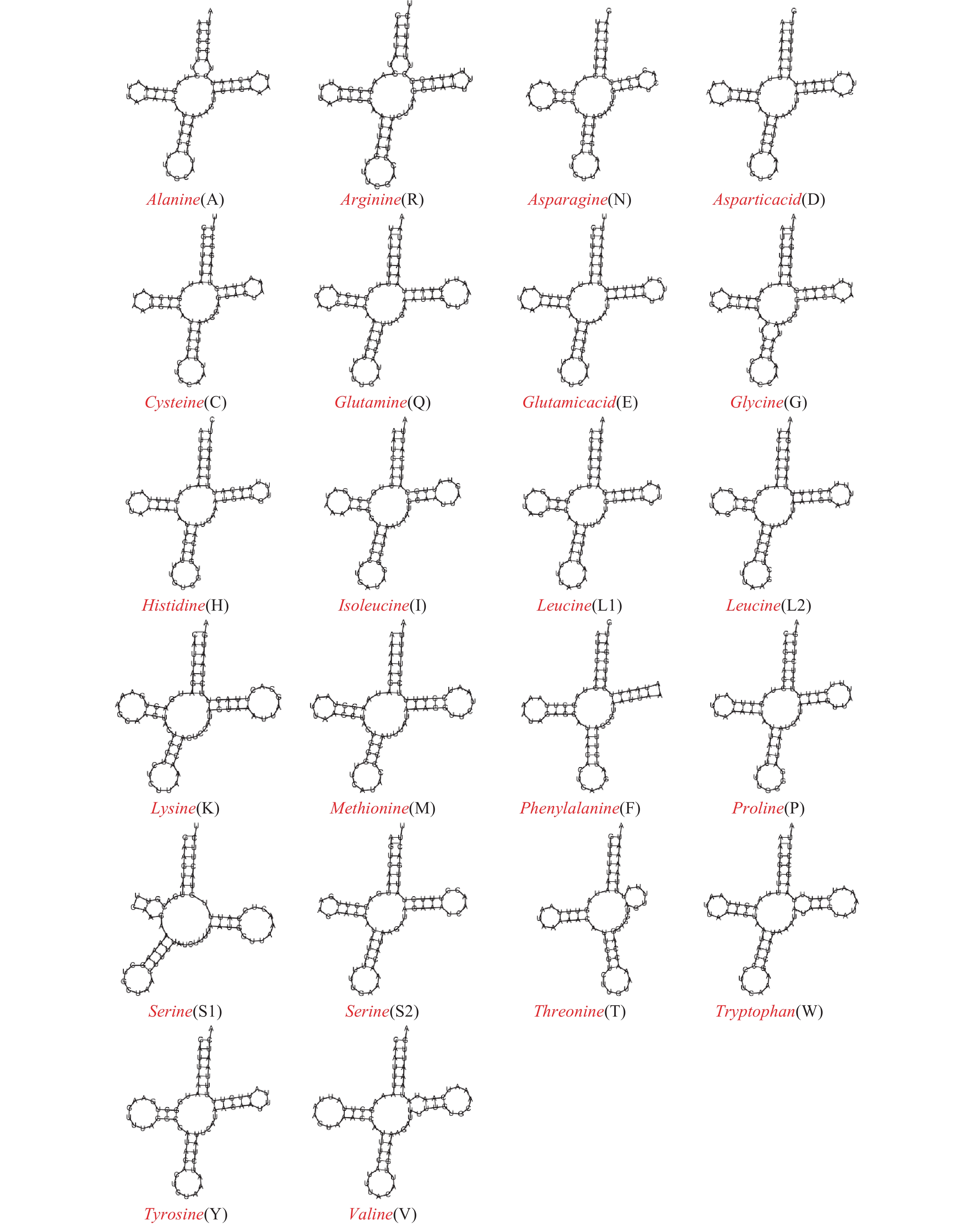
摘要:目的 明确瘤胫果实蝇[Bactrocera Bactrocera tuberculata (Bezzi) ]线粒体全基因组序列特征,探究其系统发育地位,为瘤胫果实蝇分子标记、物种鉴定和进化遗传学研究提供参考依据。方法 采用高通量测序技术对瘤胫果实蝇的线粒体全基因组进行测序、组装、拼接和注释,获得基因组全序列,并对基因组基本结构进行分析;选择NCBI 公布的果实蝇属近缘种20种实蝇的线粒体全基因组序列,基于最大似然法(Maximum likelihood, ML)进行系统发育分析。结果 瘤胫果实蝇线粒体基因组序列总长度为15 854 bp;具37个基因,其中包含13个蛋白编码基因、22个tRNA基因、2个rRNA基因和1个非编码控制基因。瘤胫果实蝇线粒体基因组全序列A+T含量为 73.2%;13个蛋白质编码基因共有密码子3 755个,在蛋白质所有22种氨基酸密码子中,UUA(leucine)的使用频率 N及相对密码子RSCU使用频率均最高,N(RSCU)为387(3.79);tRNA基因二级结构中,除苯丙氨酸(F)以及苏氨酸(T)缺少假尿嘧啶(T)环和丝氨酸(S1)缺少二氢尿嘧啶DHU环,其余19个tRNA基因均能折叠形成典型的三叶草二级结构,22个 tRNA基因中,共存在178处碱基对错配,错配碱基对均为G-U。通过对其近缘种线粒体全基因组的系统发育分析,瘤胫果实蝇与桔小实蝇(Bactrocera dorsalis)和杨桃实蝇(Bactrocera carambolae)聚在一起,表明3种实蝇的亲缘关系较近,与果实蝇亚属的其他种类聚在同个分支上,结果与形态鉴定一致。结论 首次报道了瘤胫果实蝇线粒体全基因组,GenBank登录号MW 892726,测定和分析的瘤胫果实蝇线粒体基本结构特征,核苷酸组成、系统进化分析,支持瘤胫果实蝇属于果实蝇亚属,为物种诊断、进化生物学和防治研究提供依据。Abstract:Objective Mitochondrial genome and phylogeny of Bactrocera (B.) tuberculata were studied for advancement on the molecular markers design, species identification, and evolutionary genetics relating to the destructive fruit fly.Methods High-throughput sequencing was applied to determine, assemble, join, and annotate the complete genome of B. (B.) tuberculata mitochondria. With the sequences on selected 20 species published on NCBI, the phylogeny of fruit flies was analyzed using the maximum likelihood method (ML).Results The total mitochondrial genome sequence was 15 854 bp long containing 13 protein coding genes, 22 tRNA genes, 2 rRNA genes, and one non-coding control gene with 73.2% A+T. There were 3 755 codons in the 13 protein-coding genes. Of the 22 amino acid codons in the protein, UUA (leucine) had the highest frequency (N) of 387 and relative codon (RSCU) of 3.79. Aside from the phenylalanine (F) and threonine (T) that lacked pseuduracine (T) rings and the serine (S1) without a dihydrouracil DHU ring, the secondary structures of the remaining 19 tRNA genes shaped typically like a canonical cloverleaf. And the 22 genes had 178 mismatched G-U base pairs. Based on the mitochondrial analysis, the phylogeny of B. (B.) tuberculate, B. (B.) dorsalis, and B. (B.) carambolae were closely related and in the same branch as other subgenera. The result agreed with what was revealed by the morphological observation.Conclusion For the first time, the complete mitochondrial genome of B. (B.) tuberculata was obtained with a GenBank accession number of MW 892726. The information secured on the structure and nucleotide composition of the mitochondria and the phylogenetic relation with other subgenera would aid further studies on the species identification, evolutionary biology, and pest control of the devastating pest.
-
0. 引言
【研究意义】猪流行性腹泻(Porcine epidemic diarrhea,PED)是由猪流行性腹泻病毒(Porcine epidemic diarrhea virus,PEDV)引起的一种高度接触性肠道传染病。临床上以水样腹泻、呕吐、脱水、急性肠胃炎等为主要症状[1]。各年龄阶段的猪均可感染,其中以7 日龄以内的哺乳仔猪最易感,发病也最为严重,发病率和死亡率高达100%[2]。PEDV主要通过粪便-口腔途径传播,但经粪便-鼻腔途径的空气传播在疫病的传播中也起着十分重要的作用,当受污染的粪便排入环境时便可导致大规模的流行[3],增加该病的防控难度。随着PEDV变异株的出现,该病迅速蔓延至全球[4-5],给整个养猪业造成严重的经济损失。因此,及时了解猪群的抗体保护水平,可以为PED的有效防控提供重要依据。【前人研究进展】PEDV主要侵害小肠,肠道局部黏膜免疫和特异性SIgA抗体在抗感染过程中发挥重要作用[6]。研究表明初乳或常乳中特异性SIgA抗体是保护仔猪免受PEDV攻击的最有效方式[7]。对怀孕母猪进行免疫接种成为预防和控制猪流行性腹泻、降低哺乳仔猪死亡率的关键[8]。Poonsuk等[9]研究指出IgG抗体和SIgA都有助于保护猪只免受PEDV感染。IgG分子因其分子量大,对胃酸和消化酶的抵抗力较差,而难以穿过肠壁进入肠腔发挥免疫保护功能。而SIgA抗体因其分泌片(Secretory component,SC)的存在,可以抵御胃酸和消化酶的消化作用,进入肠道后黏附在胃肠道黏膜或吸收进入血液,在呼吸道、消化道黏膜等抵御病毒、细菌等病原微生物的入侵[10]。因此,在抗PEDV感染过程,SIgA抗体发挥着重要的作用。【本研究的切入点】虽然已有学者研究了有关PEDV抗体检测的技术[11-13],但针对特异性SIgA抗体检测方法的应用较少,其相关的试剂也相对匮乏,极大阻碍了该方法的使用。SC蛋白是区分IgA和SIgA的重要标志[14]。因此,采用SIgA特异性表位的SC分泌片,能够有效区分IgA和SIgA抗体。【拟解决的关键问题】通过SC重组蛋白多克隆抗体的制备获取相关的SIgA抗体,通过纯化、标记,建立PEDV 特异性SIgA抗体间接ELISA检测方法,为PED流行病学调查及猪群免疫后抗体评估奠定基础。
1. 材料与方法
1.1 试验材料
1.1.1 毒株、试验动物和样本
猪流行性腹泻病毒(ZJ08株)由兆丰华生物科技(福州)有限公司制备、扩繁及保存;猪轮状病毒(Porcine rotavirus ,PoRV)、猪传染性胃肠炎(Porcine transmissible gastroenteritis virus,TGEV )阳性乳汁、PEDV阴性乳汁(N)和PEDV阳性乳汁(P)由本公司保存;试验大兔品种为新西兰大兔(SPF级);样本:来源于福清市某规模化猪场免疫猪流行性腹泻活疫苗和灭活疫苗的母猪群乳汁样本。
1.1.2 主要试剂
BSA购自Biosharp;HRP标记试剂盒购自Biodragon公司;TMB显色液购自博士德生物工程有限公司;ELISA酶标板购自abcam;蔗糖、PEG4000购自国药集团化学试剂有限公司;transetta(DE3)、DH5a购自Takara;6×His标签抗体购自OriGene;快速转膜液购自NCM biotech:BeyoECLMoon购自碧云天生物技术有限公司:Ni-NTA his bind resin购自上海七海复泰生物科技有限公司;HRP标记试剂盒购自Biodragon;ProteinIso®Protein G Resin购自全式金生物技术有限公司。PEDV IgA抗体检测方法(IFA法)由本实验室自建。
1.2 SC蛋白多克隆抗体的制备及纯化
1.2.1 引物设计
根据NCBI公布的猪PIgR序列(登录号:NM_214159.1)设计SC基因片段引物,并送至福州尚亚生物技术有限公司进行合成。引物序列为F:5′-CGCGGAATTCAAGAGTCC CATATTCGGTCCC-3′;R:5′-CGACAAGCTTTTCACCATCCTTCACTGCTCT-3′。
1.2.2 PEDV纯化及浓度测定
将猪流行性腹泻病毒(ZJ08株)10000 r·min−1离心处理1 h,取上清,采用PEG4000过夜沉淀处理,10000 r·min−1离心1 h,弃上清,沉淀用适量的缓冲液重悬。参照文献[15]采用蔗糖密度梯度离心的方法进行精提纯,采用RT-PCR方法和Folin-酚试剂法分别对纯化后的病毒进行PEDV鉴定及蛋白浓度测定。
1.2.3 PET32a-SC原核表达及纯化
取猪肺脏组织的总RNA为模板进行SC基因的扩增。将测序验证正确的PET32a-SC转化至transetta(DE3)中,挑选单菌落,接种至LB液体培养基中(含氨苄青霉素),37 ℃、210 r·min−1培养过夜。次日按照1∶100比例进行扩大培养,待扩增菌液OD600值达0.6时,加入一定剂量的IPTG(终浓度0.2 mmol·L−1)37 ℃继续培养5 h。收集菌液,12000 r·min−1离心20 min,弃去上清液,用预冷的PBS洗涤2次后,超声破碎处理,离心后,沉淀用8 mol·L−1脲素完全溶解,按照Ni-NTA His Bind Resin柱说明书进行重组蛋白纯化与鉴定,并采用微量分光光度计进行纯化后蛋白浓度测定。
1.2.4 Western blotting分析
将上述纯化的蛋白经12.5% SDS-PAGE电泳后,湿转法(400 mA,30 min)转印到PVDF膜上,5%脱脂奶封闭2 h,4 ℃ 6×His标签抗体孵育过夜,TBST缓冲液洗涤3次,PVDF膜置于羊抗鼠HRP-IgG抗体中,室温孵育1 h;TBST 缓冲液洗涤3次,采用ECL显色液显色并观察拍照。
1.2.5 SC蛋白多克隆抗体的制备
将5只成年健康新西兰大兔(SPF级)随机分成2组,试验组3只,对照组2只。将纯化处理的一定量的SC重组蛋白与弗氏完全佐剂按照1∶1体积比进行混合乳化后,试验组颈背部皮下注射重组SC蛋白(1 mg·只−1),并于2周后进行二免,随后每隔两周免疫1次,每次免疫剂量在前一次免疫的基础上增加20%,对照组免疫相同剂量的生理盐水。免疫6 次后,心脏采血,分离血清,−80 ℃保存备用。
1.2.6 多克隆抗体的纯化和标记
将上述采集的血清经辛酸硫酸铵方法进行粗提纯后,参照Protein G Resin说明书进行抗体纯化,按照HRP标记试剂盒说明书对纯化后的多克隆抗体进行标记。
1.3 PEDV SIgA抗体间接ELISA检测方法的建立
1.3.1 最佳反应条件的确定
采用方阵滴定法分别对PEDV抗原包被浓度(1.2、0.6、0.3、0.15 μg·孔−1)、乳汁稀释比例(1∶5、1∶10、1∶20、1∶30)、包被条件(4 ℃过夜、37 ℃ 1 h、37 ℃ 2 h)、封闭浓度(5% BSA、2% BSA、1% BSA)、封闭时间(1、2、3 h)、一抗工作条件(37 ℃ 45 min、60 min、90 min)、二抗工作条件(稀释比例1∶50、1∶100、1∶200、1∶500;作用时间37 ℃ 45、60、90 min)和TMB显色时间(10、20、25 min)等进行优化,以阳性乳汁OD450 nm/阴性乳汁OD450 nm的最大比值为最佳的反应条件。
1.3.2 临界值的确定
采用上述优化的方法,对40 份已知PEDV SIgA抗体阴性乳汁样本进行测定,计算样本中OD450 nm平均值(
¯X )和标准差(SD)。根据统计学原理,当OD450 nm≥¯X +3SD时,判定为阳性。OD450 nm≤¯X +2SD时,判定为阴性,介于两者之间的判定为可疑[16]。1.3.3 特异性试验
按照上述建立的间接ELISA方法,分别对PoRV、TGEV阳性乳汁样品进行检测,每份样品重复3次,确定该方法的特异性。
1.3.4 重复性试验
随机选取10份乳汁样本进行批内和批间间接ELISA检测,每份样品设置3个重复,计算批内和批间的变异系数,分析其重复性。
1.3.5 PEDV SIgA抗体ELISA检测方法的应用
采集52份已知免疫猪流行性腹泻疫苗的临床乳汁样本,采用上述建立的检测方法和免疫荧光方法分别对乳汁中PEDV SIgA和IgA抗体水平进行测定,比较分析IgA与SIgA抗体水平相关性。
2. 结果与分析
2.1 PEDV纯化及蛋白浓度的测定
采用PEG4000沉淀法和蔗糖密度梯度离心的方法对PEDV进行纯化,经RT-PCR结果显示,在40%~60%的蔗糖层有较高浓度的PEDV存在,其大小为327 bp(图1)。经Folin-酚试剂法对纯化样品进行蛋白浓度测定。结果显示,蛋白质量浓度为200 μg·mL−1,杂蛋白的去除率达94%。
![]() 图 1 不同浓度蔗糖层PEDV RT-PCR检测结果M:1000 DNA Marker;1~3:40%~60%层带未稀释、100倍稀释、1000倍稀释;4~6为30%层带未稀释、100倍稀释、1000倍稀释;7~9为20%层带未稀释、100倍稀释、1000倍稀释。Figure 1. RT-PCR detections of PEDV on sucrose layersM:DNA ladder DL1000; 1–3: on not diluted 40%–60% sucrose layer and on those diluted 100× and 1 000×, respectively; 4–6: on not diluted 30% sucrose layer and on those diluted 100× and 1 000×, respectively; 7–9 on not diluted 20% sucrose layer and on those diluted 100× and 1 000×, respectively.
图 1 不同浓度蔗糖层PEDV RT-PCR检测结果M:1000 DNA Marker;1~3:40%~60%层带未稀释、100倍稀释、1000倍稀释;4~6为30%层带未稀释、100倍稀释、1000倍稀释;7~9为20%层带未稀释、100倍稀释、1000倍稀释。Figure 1. RT-PCR detections of PEDV on sucrose layersM:DNA ladder DL1000; 1–3: on not diluted 40%–60% sucrose layer and on those diluted 100× and 1 000×, respectively; 4–6: on not diluted 30% sucrose layer and on those diluted 100× and 1 000×, respectively; 7–9 on not diluted 20% sucrose layer and on those diluted 100× and 1 000×, respectively.2.2 PET32a-SC原核表达蛋白的鉴定及纯化
以His标签抗体为一抗,以羊抗鼠HRP-IgG为二抗进行SDS-PAGE鉴定及Western Blot分析,结果发现在约81 kDa位置处出现目的蛋白(图2、3)。
2.3 抗原最佳包被浓度及最佳样本稀释度的确定
当抗原包被浓度为0.6 μg·孔−1,乳汁稀释比例1∶5时,P/N最大,即抗原最佳包被浓度为0.6 μg·孔−1,乳汁最佳稀释比例为1∶5(表1)。
表 1 抗原最佳包被浓度及乳汁最佳稀释度的确定(OD450 nm)Table 1. Optimal antigen concentration and milk dilution determined according to OD450 nm样本稀释度
Sample dilution抗原稀释度
Antigen dilution/(μg·孔−1)P N P/N 1∶5 1.2 0.821 0.130 6.32 0.6 0.663 0.083 7.99 0.3 0.627 0.096 6.53 0.15 0.584 0.075 7.79 1∶10 1.2 0.725 0.131 5.53 0.6 0.572 0.096 5.96 0.3 0.494 0.093 5.31 0.15 0.455 0.077 5.91 1∶20 1.2 0.69 0.132 5.23 0.6 0.563 0.105 5.36 0.3 0.472 0.115 4.10 0.15 0.436 0.099 4.40 1∶30 1.2 0.519 0.128 4.05 0.6 0.412 0.107 3.85 0.3 0.324 0.117 2.77 0.15 0.271 0.111 2.44 2.4 最佳抗原包被条件的确定
间接ELISA检测结果显示,当抗原37 ℃作用2 h后,P/N最大值达7.65。因此确定抗原最佳包被条件为37 ℃ 2 h(表2)。
表 2 最佳包被条件的确定Table 2. Optimum conditions for coating指标 Index 包被条件 Coating condition 4 ℃包被过夜
4 ℃ wrap overnight37 ℃ 1 h 37 ℃ 2 h P 0.657 0.572 0.620 N 0.130 0.082 0.081 P/N 5.05 6.98 7.65 2.5 最佳封闭条件的确定
间接ELISA结果显示,用2% BSA封闭液、 37 ℃条件下封闭2 h的P/N有最大值。因此确定最佳的封闭条件为:用2% BSA,37 ℃作用2 h(表3)。
表 3 最佳封闭条件的确定Table 3. Optimum conditions for blocking指标 Index 封闭浓度
Closure condition封闭条件(37 ℃)
Closure condition1% 2% 5% 1 h 2 h 3 h P 1.180 1.121 1.034 1.110 1.070 1.077 N 0.241 0.213 0.220 0.282 0.226 0.255 P/N 4.89 5.26 4.68 3.94 4.73 4.22 2.6 最佳乳汁工作条件的确定
采用不同的孵育时间进行ELISA检测,结果显示,将乳汁样本置37 ℃作用60 min时,P/N最大,因此乳汁的最佳工作条件为37 ℃孵育60 min(表4)。
表 4 最佳乳汁样本工作条件的确定Table 4. Optimum working conditions for milk specimens指标
Index孵育时间 Incubation time 37 ℃ 45 min 37 ℃ 60 min 37 ℃ 90 min P 0.638 0.639 0.654 N 0.096 0.085 0.087 P/N 6.65 7.52 7.52 2.7 最佳标记抗体工作浓度的确定
对上述标记的酶标抗体进行不同比例的稀释后,采用上述建立的ELISA进行检测,标记抗体的稀释比例为1∶100、37 ℃作用90 min时,P/N值最大。因此,标记抗体的最佳稀释比例为1∶100倍,37 ℃孵育90 min(表5)。
表 5 最佳标记抗体工作条件的确定Table 5. Optimum working conditions for HRP-labeled secondary antibody指标
Index稀释比例
dilution作用时间
Reaction conditions1∶50 1∶100 1∶200 1∶300 37 ℃ 45 min 37 ℃ 60 min 37 ℃ 90 min P 0.819 0.891 0.709 0.706 0.746 0.796 0.867 N 0.168 0.162 0.172 0.159 0.146 0.150 0.160 P/N 4.88 5.5 4.12 4.44 5.11 5.31 5.42 2.8 底物最佳反应时间的确定
采用不同的TMB作用时间进行ELISA检测,结果发现,TMB反应时间为20 min时,P/N值最大。因此,底物最佳反应时间为20 min(表6)。
表 6 底物最佳反应时间的确定Table 6. Optimum reaction time for TMB指标 Index 孵育时间 Incubation time 15 min 20 min 25 min 30 min P 0.869 0.934 1.019 1.089 N 0.136 0.136 0.215 0.187 P/N 6.39 6.87 4.93 5.82 2.9 临界值判定
采用上述方法对40份经IFA检测的阴性乳汁样本进行检测,结果显示:40份阴性乳汁样本的平均值(
¯X )为0.232,标准差(SD)为0.056;根据统计学原理,确定阴阳性临界值为P=¯X +3SD=0.4,N=¯X +2SD=0.34。当待测样本OD450 nm值≥0.4判定为阳性;当待测样本OD450 nm值≤0.34判定为阴性;介于两者之间判定为可疑。2.10 特异性试验
按照上述建立的间接ELISA方法,分别对PoRV、TGEV阳性乳汁样品进行检测。结果表明,本研究建立的间接ELISA方法与上述乳汁样本均无交叉反应(OD450 nm<0.10),具有良好的特异性。
2.11 重复性试验
采用上述建立的间接ELISA方法对10份乳汁样本进行批内和批间重复性检测,结果显示,批内变异系数在0.3%~1.3%,批间变异系数在1%~6%,表明该方法具有很好的重复性(表7)。
表 7 乳汁样本的重复性检验结果Table 7. Repeatability of whey samples编号
Number组内重复
Intra group repetition组间重复
Intergroup duplication¯X±SD 变异系数CV/% ¯X±SD 变异系数CV/% 1 0.167±0.009 0.3 0.168±0.040 1 2 0.225±0.005 0.2 0.223±0.052 2 3 0.249±0.006 0.2 0.252±0.009 1 4 0.352±0.017 0.6 0.342±0.032 2 5 0.304±0.006 0.2 0.298±0.028 1 6 0.340±0.009 0.3 0.335±0.025 2 7 0.159±0.040 1.3 0.162±0.059 3 8 0.641±0.002 0.1 0.623±0.066 6 9 0.174±0.021 0.8 0.165±0.032 5 10 0.161±0.013 0.5 0.157±0.035 1 2.12 PEDV SIgA抗体检测方法的应用
采用上述建立的ELISA检测方法对临床采集的乳汁样本进行测定,并通过免疫荧光方法对PEDV IgA抗体进行测定。结果显示,52份样本中PEDV IgA抗体阳性率达98.1%,而PEDV SIgA抗体阳性率仅为69.2%。从样品的符合率情况分析发现,PEDV IgA抗体为阳性的样本,其特异性的SIgA抗体阳性率仅为70.6%,提示乳汁中IgA抗体的测定虽然对疫苗的免疫情况存在一定的指导意义,但要真实了解猪群的实际免疫保护情况还需对特异性的SIgA抗体水平进行测定。
3. 讨论与分析
PEDV作为一种肠道性传染病,其特异性SIgA抗体能够抑制PEDV在肠道上皮细胞的附着与增殖,保护仔猪肠道上皮细胞,阻止PEDV侵入机体[17]。目前已经有很多学者针对PEDV特异性抗体的ELISA检测方法进行探讨[18]。但其大多采用中和表位的部分蛋白作为抗原[19],或者采用针对IgA抗体的检测方法[20,21]。虽然也有学者采用PEDV抗原以及SIgA单克隆抗体制备等方法建立了PEDV特异性SIgA抗体检测方法,但因单克隆细胞制备存在筛选难度大、过程繁琐、培养环境条件苛刻等,很难推广应用。
采用本研究建立的方法与免疫荧光方法对PEDV特异性IgA抗体和SIgA特异性抗体检测结果比较分析发现,样品中IgA抗体为阳性时,其特异性的SIgA抗体不一定呈现阳性;样品中IgA抗体阴性的样本,其特异性SIgA抗体基本呈现阴性。这一发现可能是当前猪群免疫疫苗后,乳汁中IgA抗体水平高但仍然出现仔猪拉稀这一现象的原因,也进一步说明了通过乳汁中特异性IgA抗体水平的测定不能全面反映PEDV SIgA抗体的水平情况。由于初乳中不仅存在SIgA,也存在IgA。仔猪吮吸母乳后,其单体IgA通常经过血液循环发挥作用,而SIgA则可以锚定在肠道黏膜表面形成保护层。因此,通过测定乳汁中IgA抗体水平反映的可能是乳汁中的总IgA水平,这也是导致临床样本中PEDV IgA抗体为阳性,其特异性的SIgA抗体为阴性的可能原因。
本研究采用PET32a-SC重组蛋白作为免疫原制备SIgA特异性多克隆抗体,通过对获取的多克隆抗体进行纯化及辣根过氧化物酶标记后,建立了能够特异性识别SIgA抗体的诊断方法。与柏家果等[22]建立的间接ELISA方法相比,本研究通过全病毒作为包被抗原,大大提高了该方法的特异性和灵敏度。以标记的多克隆抗体作为酶标抗体,简化了HRP标记的山羊抗体IgG作为酶标三抗的操作过程,大大提高了工作效率,降低了操作强度。本研究方法的建立为猪群免疫猪流行性腹泻病毒相关疫苗后提供了便捷、有效的检验方法,为PEDV特异性黏膜免疫SIgA抗体水平的监测提供技术手段。因缺乏相关的特异性SIgA抗体检测试剂,PEDV SIgA抗体水平与动物免疫保护之间的关系还有待于结合猪群临床表现及免疫情况进行进一步验证。
-
表 1 瘤胫果实蝇线粒体基因组特征
Table 1 Characteristics of mitochondrial genome of B. (B.) tuberculata
基因
Gene位置
Location/bp基因长度
Gene length/bp编码链
Strand
J(+)、N(-)基因间隔
Intergenic
sequence/bp起始密码子
Start
codon终止密码子
Stop
codontrnI-GAU 1~66 66 J trnQ-UUG 64~132 69 N −3 trnM-CAU 201~269 69 J 68 ND2 270~1292 1023 J 0 ATT TAA trnW-UCA 1303~1371 69 J 10 trnC-GCA 1364~1426 63 N −8 trnY-GUA 1473~1539 67 N 46 COX1 1538~3076 1539 J −2 TCG TAA trnL-UAA 3072~3137 66 J −5 COX2 3142~3831 690 J 4 ATG TAA trnK-CUU 3836~3906 71 J 4 trnD-GUC 3910~3976 67 J 2 ATP8 3977~4138 162 J 0 GTG TAA ATP6 4132~4809 678 J −7 ATG TAA COX3 4809~5597 789 J −1 ATG TAA trnG-UCC 5607~5671 65 J 9 ND3 5672~6025 354 J 0 ATT TAG trnA-UGC 6024~6088 65 J −2 trnR-UCG 6096~6159 64 J 7 trnN-GUU 6171~6235 65 J 11 trnS-GCU 6236~6303 68 J 0 trnE-UUC 6304~6370 67 J 0 trnF-GAA 6392~6456 65 N 21 ND5 6437~8176 1740 N −20 ATT TAG trnH-GUG 8192~8257 66 N 15 ND4 8258~9598 1341 N 0 ATG TAG ND4L 9592~9888 297 N −7 ATG TAA trnT-UGU 9891~9950 60 J −8 trnP-UGG 9971~10036 66 N 20 ND6 10039~10563 525 J 2 ATT TAA CYTB 10563~11697 1135 J −1 ATG ATT trnS-UGA 11697~12725 1029 N −1 ATA TAA ND1 11704~11770 67 J −22 trnL-UAG 12736~12800 65 N 965 l-rRNA 12778~14135 1358 N −22 trnV-UAC 14133~14204 72 N −3 s-rRNA 14204~14995 791 N −1 控制区
Control region14996~15943 947 J 0 表 2 瘤胫果实蝇线粒体全基因组碱基组成
Table 2 Bases in mitochondrial genome of B. (B.) tuberculata
区域
RegionA/% C/% G/% T/% A+T/% AT偏移
AT SkewGC偏移
GC Skew基因长度
Gene length/bp全基因组
Whole genome39.1 16.4 10.4 34.1 73.2 0.068 −0.224 15943 PCGs 29.9 14.9 14.4 40.8 70.7 −0.154 −0.017 11302 PCGs J 32.0 18.7 12.7 36.6 68.6 −0.067 −0.191 6895 PCGs N 26.7 9.0 17.0 47.2 73.9 −0.277 0.308 4407 ND2 34.1 16.8 9.1 40.0 74.1 −0.080 −0.297 1023 COXI 30.1 18.9 16.2 34.8 64.9 −0.072 −0.077 1539 COX2 33.5 18.4 14.1 34.1 67.6 −0.009 −0.132 690 ATP8 37.7 17.9 8.6 35.8 73.5 0.026 −0.351 162 ATP6 30.4 19.9 11.8 37.9 68.3 −0.110 −0.256 678 COX3 29.7 19.4 15.3 35.6 65.3 −0.090 −0.118 789 ND3 31.9 16.7 9.9 41.5 73.4 −0.131 −0.256 354 ND5 27.8 9.0 17.1 46.0 73.8 −0.247 0.310 1740 ND4 26.3 8.9 16.9 47.8 74.1 −0.290 0.310 1341 ND4L 26.6 7.1 14.5 51.9 78.5 −0.322 0.343 297 ND6 37.0 16.6 6.9 39.6 76.6 −0.034 −0.413 525 CYTB 30.9 20.7 13.5 34.9 65.8 −0.061 −0.211 1135 ND1 25.5 9.7 17.6 47.2 72.7 −0.298 0.289 1029 tRNA 36.9 11.1 14.1 37.8 74.7 −0.012 0.119 1462 tRNA J 37.2 12.7 13.2 36.8 74.0 0.005 0.019 929 tRNA N 36.4 8.4 15.6 39.6 76.0 −0.042 0.300 533 rRNA 35.7 8.0 14.6 41.7 77.4 −0.078 0.292 2149 1-rRNA 36.7 7.1 13.8 42.5 79.2 −0.073 0.321 1358 s-rRNA 34.1 9.5 16.1 40.3 74.4 −0.083 0.258 791 控制区
Control region46.9 6.6 5.0 41.5 88.4 0.061 0.138 947 表 3 瘤胫果实蝇线粒体基因组蛋白质编码基因密码子使用统计
Table 3 Statistics of codon usage in mitochondria of B. (B.) tuberculata
密码子 Codon N R 密码子 Codon N R 密码子 Codon N R 密码子 Codon N R UUU(F) 235 1.42 UCU (S) 114 2.68 UAU(Y) 144 1.73 UGU(C) 41 1.91 UUC(F) 96 0.58 UCC (S) 10 0.24 UAC(Y) 22 0.27 UGC(C) 2 0.9 UUA(L) 387 3.79 UCA (S) 95 2.24 UAA(*) 0 0.00 UGA(W) 95 1.88 UUG(L) 68 0.67 UCG (S) 2 0.05 UAG(*) 0 0.00 UGG(W) 6 0.12 CUU(L) 66 0.65 CCU(P) 75 2.17 CAU(H) 55 1.39 CGU(R) 15 1.02 CUC(L) 5 0.05 CCC(P) 13 0.38 CAC(H) 24 0.61 CGC(R) 3 0.20 CUA(L) 81 0.79 CCA(P) 48 1.39 CAA(Q) 74 1.87 CGA(R) 37 2.51 CUG(L) 5 0.05 CCG(P) 2 0.06 CAG(Q) 5 0.13 CGG(R) 4 0.27 AUU(I) 298 1.76 ACU (T) 79 1.57 AAU(N) 163 1.72 AGU(S) 47 1.11 AUC(I) 40 0.24 ACC (T) 19 0.38 AAC(N) 26 0.28 AGC(S) 9 0.21 AUA(M) 174 1.73 ACA (T) 101 2.01 AAA(K) 63 1.42 AGA(S) 63 1.48 AUG(M) 27 0.27 ACG (T) 2 0.04 AAG(K) 26 0.58 AGG(S) 0 0.00 GUU(V) 84 1.53 GCU(A) 116 2.43 GAU(D) 58 1.63 GGU(G) 71 1.23 GUC(V) 8 0.15 GCC(A) 22 0.46 GAC(D) 13 0.37 GGC(G) 3 0.05 GUA(V) 112 2.02 GCA(A) 50 1.05 GAA(E) 73 1.95 GGA(G) 126 2.18 GUG(V) 16 0.29 GCG(A) 3 0.06 GAG(E) 2 0.05 GGG(G) 31 0.54 N为线粒体全基因组蛋白质编码基因的密码子使用个数;R:RSCU为相对同义密码子的使用频率。*:终止密码子。
N: number of codons used for protein-coding genes of whole mitochondrial genome; R: frequency of use of relative synonymous codon (RSCU). *: stop codon. -
[1] 邓裕亮, 李正跃, 蒋小龙, 等. 缅甸第四特区实蝇类害虫调查 [J]. 植物检疫, 2005, 19(5):316−318. DENG Y L, LI Z Y, JIANG X L, et al. Investigation on fruit fly pests in the fourth special zone of Myanmar [J]. Plant Quarantine, 2005, 19(5): 316−318. (in Chinese)
[2] 邓裕亮, 李志红, 白永华, 等. 老挝中北部地区果实蝇属(Bactrocera)害虫种类初步调查 [J]. 植物检疫, 2010, 24(1):52−53. DOI: 10.3969/j.issn.1005-2755.2010.01.021 DENG Y L, LI Z H, BAI Y H, et al. Preliminary investigation on insect species of Bactrocera in central and northern Laos [J]. Plant Quarantine, 2010, 24(1): 52−53. (in Chinese) DOI: 10.3969/j.issn.1005-2755.2010.01.021
[3] DREW R A I, ROMIG M C, DORJI C. Records of Dacine fruit flies and new species of Dacus (Diptera: Tephritidae) in Bhutan [J]. The Raffles Bulletin of Zoology, 2007, 55(1): 1−21.
[4] LEBLANC L, HOSSAIN M A, KHAN S, et al. Additions to the Fruit Fly Fauna (Diptera: Tephritidae: Dacinae) of Bangladesh, with a Key to the Species [J]. Proceedings of the Hawaiian Entomological Society, 2014, 46: 31−40.
[5] 蒋小龙. 云南边境检疫性实蝇风险分析研究 [J]. 西南农业大学学报, 2002, 24(5):402−405,421. DOI: 10.3969/j.issn.1673-9868.2002.05.006 JIANG X L. Risk analysis of quarantine fruit flies along Yunnan borber [J]. Journal of Southwest Agricultural University, 2002, 24(5): 402−405,421. (in Chinese) DOI: 10.3969/j.issn.1673-9868.2002.05.006
[6] 黄振, 侯有明, 郭琼霞, 等. 利用物种特异性PCR技术快速鉴定南瓜实蝇 [J]. 福建农业学报, 2021, 36(2):215−220. HUANG Z, HOU Y M, GUO Q X, et al. Rapid species-specific PCR identification of Bactrocera tau(Diptera: Tephritidae) [J]. Fujian Journal of Agricultural Sciences, 2021, 36(2): 215−220. (in Chinese)
[7] BAE J S, KIM I, SOHN H D, et al. The mitochondrial genome of the firefly, Pyrocoelia rufa: Complete DNA sequence, genome organization, and phylogenetic analysis with other insects [J]. Molecular Phylogenetics and Evolution, 2004, 32(3): 978−985. DOI: 10.1016/j.ympev.2004.03.009
[8] 徐浪, 余道坚, 张润杰, 等. 桔小实蝇线粒体基因组全序列及其分析 [J]. 昆虫学报, 2007, 50(8):755−761. DOI: 10.3321/j.issn:0454-6296.2007.08.001 XU L, YU D J, ZHANG R J, et al. Sequencing and analysis of the complete mitochondrial genome of the Oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) [J]. Acta Entomologica Sinica, 2007, 50(8): 755−761. (in Chinese) DOI: 10.3321/j.issn:0454-6296.2007.08.001
[9] 叶军, 房蕊, 易建平, 等. 4种实蝇线粒体DNA全序列测定 [J]. 植物检疫, 2010, 24(3):11−14. DOI: 10.3969/j.issn.1005-2755.2010.03.004 YE J, FANG R, YI J P, et al. The complete sequence determination and analysis of four species of bactrocera mitochondrial genome [J]. Plant Quarantine, 2010, 24(3): 11−14. (in Chinese) DOI: 10.3969/j.issn.1005-2755.2010.03.004
[10] 梅琰, 岳巧云, 贾凤龙. 双翅目昆虫线粒体基因组研究进展 [J]. 环境昆虫学报, 2012, 34(4):497−503. DOI: 10.3969/j.issn.1674-0858.2012.04.15 MEI Y, YUE Q Y, JIA F L. Research progress on mitochondrial genomes of Dipteral insect [J]. Journal of Environmental Entomology, 2012, 34(4): 497−503. (in Chinese) DOI: 10.3969/j.issn.1674-0858.2012.04.15
[11] 姜帆, 李志红, 梁亮, 等. 实蝇科昆虫线粒体基因组研究进展 [J]. 植物检疫, 2016, 30(3):12−17. JIANG F, LI Z H, LIANG L, et al. Research progress in mitochondrial genomes of Tephritidae insect [J]. Plant Quarantine, 2016, 30(3): 12−17. (in Chinese)
[12] 梁亮, 江威, 余慧, 等. 中国果实蝇属种类的DNA条形码鉴定(双翅目, 实蝇科)[J]. 动物分类学报, 2011, 36(4): 925−932. LIANG L, JIANG W, YU H, et al. Identification of Chinese bactrocera species through DNA barcoding (Diptera, Tephritidae)[J]. Acta Zootaxonomica Sinica, 2011, 36(4): 925−932. (in Chinese)
[13] 范京安, 顾海丰, 陈世界, 等. 实蝇科昆虫mtDNA-COI基因序列分析及系统发育研究[J]. 西南大学学报: 自然科学版, 2009, 31(10): 88-95. FAN J A, GU H F, CHEN S J, et al. Sequence analysis of mtDNA-COI gene and molecular phylogeny of Fruitfly(Tepbritidae)[J]. Journal of Southwest University (Natural Science Edition), 2009, 31(10): 88-95. (in Chinese)
[14] 刘慎思, 张桂芬, 万方浩. 基于mtDNA COI基因的离腹寡毛实蝇属常见种DNA条形码识别和系统发育分析 [J]. 昆虫学报, 2014, 57(3):343−355. LIU S S, ZHANG G F, WAN F H. DNA barcoding and phylogenetic analysis of common species of the genus Bactrocera(Diptera: Tephritidae) based on mtDNA COI gene sequences [J]. Acta Entomologica Sinica, 2014, 57(3): 343−355. (in Chinese)
[15] KROSCH M N, SCHUTZE M K, ARMSTRONG K F, et al. A molecular phylogeny for the Tribe Dacini (Diptera: Tephritidae): Systematic and biogeographic implications [J]. Molecular Phylogenetics and Evolution, 2012, 64(3): 513−523. DOI: 10.1016/j.ympev.2012.05.006
[16] CLARY D O, WOLSTENHOLME D R. The mitochondrial DNA molecule ofDrosophila yakuba: Nucleotide sequence, gene organization, and genetic code [J]. Journal of Molecular Evolution, 1985, 22(3): 252−271. DOI: 10.1007/BF02099755
[17] BOORE J L. Animal mitochondrial genomes [J]. Nucleic Acids Research, 1999, 27(8): 1767−1780. DOI: 10.1093/nar/27.8.1767
[18] CLARY D O, WOLSTENHOLME D R. Drosophila mitochondrial DNA: Conserved sequences in the A+T-rich region and supporting evidence for a secondary structure model of the small ribosomal RNA [J]. Journal of Molecular Evolution, 1987, 25(2): 116−125. DOI: 10.1007/BF02101753
[19] 林兴雨, 刘向阳, 宋南. 大赤隐翅虫的线粒体基因组研究及系统发育分析 [J]. 福建农业学报, 2023, 38(5):607−615. LIN X Y, LIU X Y, SONG N. Mitochondrial genome and phylogenetics of Philonthus spinipes(Sharp, 1874) [J]. Fujian Journal of Agricultural Sciences, 2023, 38(5): 607−615. (in Chinese)
[20] BOORE J L, LAVROV D V, BROWN W M. Gene translocation links insects and crustaceans [J]. Nature, 1998, 392(6677): 667−668. DOI: 10.1038/33577
[21] 王菊平, 宣善滨, 张育平, 等. 武铠蛱蝶线粒体基因组全序列测定和分析 [J]. 昆虫学报, 2015, 58(3):319−328. WANG J P, XUAN S B, ZHANG Y P, et al. Sequencing and analysis of the complete mitochondrial genome of Chitoria ulupi(Lepidoptera: Nymphalidae) [J]. Acta Entomologica Sinica, 2015, 58(3): 319−328. (in Chinese)
[22] ZHANG Y, FENG S Q, ZENG Y Y, et al. The first complete mitochondrial genome of Bactrocera tsuneonis (Miyake) (Diptera: Tephritidae) by next-generation sequencing and its phylogenetic implications[J]. International Journal of Biological Macromolecules, 2018, 118(Pt A): 1229-1237.
[23] 黄振, 郭琼霞. 基于种特异性SS-COⅠ技术快速鉴定瘤胫实蝇 [J]. 生物安全学报, 2022, 31(2):179−184. DOI: 10.3969/j.issn.2095-1787.2022.02.013 HUANG Z, GUO Q X. Rapid identification of Bactrocera tuberculata (Diptera: Tephritidae) by species-specific SS-COⅠ technique [J]. Journal of Biosafety, 2022, 31(2): 179−184. (in Chinese) DOI: 10.3969/j.issn.2095-1787.2022.02.013
[24] ISAZA J P, ALZATE J F, CANAL N A. Complete mitochondrial genome of the Andean morphotype of Anastrepha fraterculus (Wiedemann) (Diptera: Tephritidae) [J]. Mitochondrial DNA Part B, Resources, 2017, 2(1): 210−211. DOI: 10.1080/23802359.2017.1307706
[25] 周志军, 杨明茹, 常岩林, 等. 两种纺织娘线粒体基因组的比较分析 [J]. 昆虫学报, 2013, 56(4):408−418. ZHOU Z J, YANG M R, CHANG Y L, et al. Comparative analysis of mitochondrial genomes of two long-legged katydids (Orthoptera: Tettigoniidae) [J]. Acta Entomologica Sinica, 2013, 56(4): 408−418. (in Chinese)
-
期刊类型引用(0)
其他类型引用(1)








 下载:
下载: