SNP Markers Development and Genetic Relationship Identification of Germplasm Resources of Canarium album Based on SLAF-Seq Technology
-
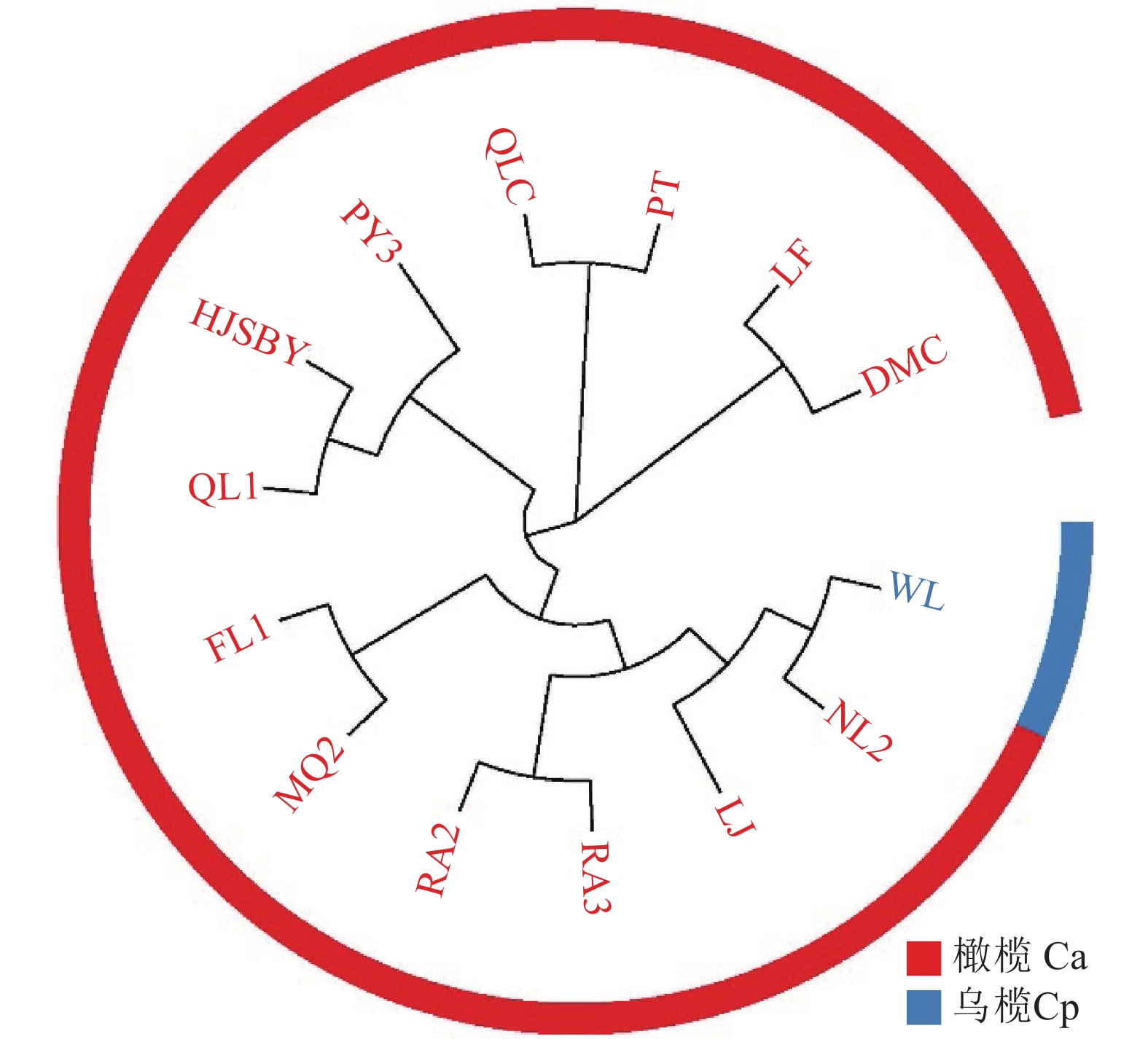
摘要:目的 开发橄榄SNP标记并分析其种质资源遗传多样性,为橄榄种质资源保护和利用提供依据。方法 基于SLAF-Seq技术进行橄榄SNP标记开发,同时采用系统进化分析、群体聚类分析和主成分分析等研究了橄榄种质资源遗传结构和遗传多样性。结果 基于SLAF-Seq技术共挖掘到506 701个SLAF标签,其中多态性SLAF标签27 108个,开发获得361 386个群体SNP标记;基于SNP标记,利用系统进化树和群体聚类分析可分别将橄榄种质资源分为3和6个类群,整体Nei多样性指数和Shanon-Wiener指数分别为0.321和0.472。两种分类方法分析结果均发现,不同地区之间的橄榄种质资源并未严格按照地域分布归类。结论 橄榄种质资源遗传多样性相对丰富,且不同地域间存在种质资源交流,而采用SNP标记可有效鉴定橄榄种质资源。Abstract:Objective SNP markers development and genetic diversity analysis of Canarium album were perfomed to facilitate the protection and utilization of germplasm resources of C. album.Method SNP markers of C. album were developed based on SLAF-Seq technology, and the genetic structure and genetic diversity were studied by phylogenetic analysis, population cluster analysis and principal component analysis.Result A total of 506 701 SLAF loci were mined, including 27 108 polymorphic SLAF loci, and 361 386 SNP markers were developed. Based on these SNP markers, germplasm resources of C. album were respectively divided into 3 and 6 groups by phylogenetic tree and population cluster analysis. The overall Nei diversity index and Shanon Wiener index were 0.321 and 0.472, respectively. The results of two classification methods showed that germplasm resources of C. album from different regions were not classified strictly according to regional distribution.Conclusion The genetic diversity of germplasm resources of C. album had great genetic diversity which were exchange frequently among different regions, and the developed SNP markers of C. album could effectively identify the germplasm resources.
-
Keywords:
- Canarium album /
- germplasm resource /
- SNP marker /
- genetic structure /
- genetic diversity
-
羊的传统放牧方式受到天然草场营养物质的季节性不平衡、载畜量有限以及草地生态环境恶化等因素的制约[1],舍饲圈养是我国未来肉羊饲养方式的发展方向[2-3]。有研究者比较了不同饲养方式对南江黄羊[3]、浏阳黑山羊[4]等肉羊品种生长发育的影响。福清山羊是福建省优良地方肉用品种,具有早熟、繁殖力高、耐粗饲、肉质风味好等优点,深受消费者喜爱[5]。谢喜平等研究了福清山羊和成都麻羊F1代早期生长发育规律[6],李文杨等采用生长曲线拟合法研究了放牧福清山羊体重生长发育规律[7],但尚未见不同饲养方式对福清山羊生长发育影响的报道。为了深入保护和开发福清山羊种质资源,拟比较放牧和舍饲圈养模式下福清山羊羯羊的生长发育规律,并利用Von Bertalalanffy、Gompertz和Logistic非线性生长模型对不同饲养方式的福清山羊羯羊体重增长进行拟合分析,为福清山羊商品羊的饲养管理和生产模式优化提供理论依据。
1. 材料与方法
1.1 试验材料与饲养管理
试验在福建省农业科学院福清市渔溪肉羊试验基地进行。采用单因素试验设计,选择体重、体况相近的健康3月龄断奶福清山羊(2017年1-2月出生)羯羊90只,平均体重(9.33±1.34) kg。按照饲养方式分为全圈养组(A组)、圈养+放牧组(B组)、放牧组(C组)3组,每组30只,各组在相同面积的羊栏(14.0 m×6.3 m)内单栏饲养。其中A组不放牧(全圈养饲养),B组和C组在非雨天进行每天4~7 h的相同时长放牧,B组的放牧地为固定的15 m×30 m舍外草地(圈养结合放牧饲养),C组天然草场人工放牧(放牧饲养)。试验开始前对试验羊只进行体重、体尺测定和统一编号。
试验日粮参考肉羊饲养标准(NY/T816-2004)(日增重为100 g·d-1)按照4~6月龄(前期)、7~9月龄(中期)、10~12月龄(后期)3个时期分别设计。A组、B组饲喂日粮、日饲喂量相同,前、中、后期饲喂量(干物质计)分别为0.56、0.61、0.66 kg·d-1;C组放牧后进行补饲,补饲日粮和补饲量与A组日粮中精料配方一致;早晚各饲喂(补饲)1次。不同时期各日粮组成及营养水平见表 1。试验期间各组羊只自由采食和饮水,栏舍环境条件及其他饲养管理均相同。
表 1 试验日粮组成及营养水平(干物质基础)Table 1. Formulation and nutritional composition of experimental diet (DM basis)项目 4~6月龄 7~9月龄 10~12月龄 A、B组 C组 A、B组 C组 A、B组 C组 精料 牧草 精料 牧草 精料 牧草 原料 天然牧草 100 100 100 玉米青贮/% 30 30 30 杂交狼尾草/% 30 30 30 玉米/% 18 45 25 62.5 23 57.5 大豆粕/% 10.3 25.75 4 10 2.5 6.25 麸皮/% 8.2 20.5 7.4 18.5 11 27.5 磷酸氢钙/% 1.1 2.75 1.2 3 1.15 2.875 食盐/% 0.5 1.25 0.5 1.25 0.5 1.25 小苏打/% 0.5 1.25 0.55 1.375 0.5 1.25 石粉/% 0.9 2.25 0.85 2.125 0.85 2.125 预混料①/% 0.5 1.25 0.5 1.25 0.5 1.25 合计/% 100 100 100 100 100 100 100 100 100 营养水平 消化能②/(MJ·kg-1) 6.39 12.52 6.39 12.50 6.33 12.35 粗蛋白/% 13.21 18.02 7.57 10.67 12.45 10.24 10.44 11.75 6.14 钙/% 0.86 1.79 0.98 0.79 1.72 0.95 0.74 1.66 1.02 磷/% 0.57 1.02 0.09 0.49 1.08 0.18 0.50 1.11 0.11 中性洗涤纤维/% 47.28 29.11 66.77 52.77 37.69 61.33 56.71 38.11 67.48 酸性洗涤纤维/% 27.67 11.75 41.97 29.61 13.94 39.66 31.31 13.46 42.15 注:①每千克预混料中含有:VA 200000 IU,VD 50000 IU,VE 500 IU,Fe 2 g,Cu 0.75 g,Zn 3 g,Mn 4 g,I 50 mg,Se 20 mg,Co 50 mg。②除消化能为计算值外,其他均为实测值。 1.2 体重测定
试验全期9个月,于每个时期的前1 d以及试验结束当天上午逐只空腹称重。
1.3 拟合分析模型
选用Von Bertalanffy、Gompertz和Logistic 3种数学模型作为非线性生长模型,对不同饲养方式福清山羊体重生长过程进行拟合。各模型中W表示t月龄时的体重估计值,参数A为成熟体重,K为瞬时相对生长率,B为调节参数,t为月龄(表 2)。
表 2 拟合分析的3种非线性生长模型表达式及特征参数Table 2. Three nonlinear models and parameters for analyzing growth of Fuqing goats模型 表达式 拐点体重 拐点月龄 Von Bertalanffy W = A(1- Be-Kt)3 8A/27 (ln3B)/K Gompertz W = Ae-bexp(-Kt) A/e (lnB)/K Logistic W = A/(1+Be-Kt) A/2 (lnB)/K 1.4 数据处理及分析方法
参考戴国俊等[8]的方法,利用SPSS17.0软件进行Von Bertalanffy、Gompertz和Logistic 3种非线性生长模型分析,并估计各模型的A、B、
K参数和拟合度R2,进一步建立各生长曲线模型。基础数据统计计算采用Excel 2007。 2. 结果与分析
2.1 不同饲养方式下福清山羊生长情况
不同饲养方式下,相同月龄放牧组试验羊的体重、平均日增重和相对生长率均为最低(表 3)。3种饲养方式试验羊的6月龄体重差异不显著(P>0.05),A组体重最大,为15.83 kg。B组试验羊9月龄和12月龄的体重显著高于放牧组(P<0.05),分别为20.95和26.03 kg;A组2个阶段的体重高于C组,二者差异不显著水平(P>0.05)。
表 3 不同饲养方式福清山羊体重增长的影响Table 3. Effect of husbandry methods on body weight of Fuqing goats月龄 饲养方式 体重/kg 平均日增重/(g·d-1) 相对生长率/% 3 A组 9.32±1.37 - - B组 9.33±1.35 - - C组 9.34±1.35 - - 6 A组 15.98±1.79 73.96±6.40a 53.00±4.20a B组 15.83±1.81 72.30±5.90a 52.05±3.35a C组 15.39±1.81 67.30±5.82b 49.30±3.40b 9 A组 20.67±2.88ab 51.19±12.61b 25.28±3.26b B组 20.95±2.09a 56.89±5.74a 27.99±3.01a C组 19.60±2.23b 46.70±6.23c 24.07±2.13b 12 A组 25.09±4.60ab 49.04±20.51ab 18.48±6.10ab B组 26.03±3.10a 56.44±13.74a 21.35±4.36a C组 23.62±3.81b 44.67±19.38b 17.89±6.54b 注:同列数据后不同小写字母表示相同月龄之间差异显著(P<0.05)。 随着日龄的增长,各饲养方式的平均日增重和相对生长率均呈下降趋势(表 3),符合畜禽体重生长发育的一般规律。整个试验周期内,3~6月龄福清山羊羯羊的生长速度最快,其中A组平均日增重和相对生长率最大,分别为73.96 g·d-1、53.00%,A组和B组平均日增重和相对生长率显著高于C组(P<0.05)。6~12月龄,B组平均日增重和相对生长率最大,均显著高于C组(P<0.05),6~9月龄时显著高于A组(P<0.05),9~12月龄与A组差异不显著;C组全期平均日增重和相对生长率均最小。
2.2 不同饲养方式下福清山羊生长模型拟合分析
不同饲养方式下福清山羊生长模型拟合结果见表 4。拟合度(R2)是评价非线性生长模型拟合实际生长发育的主要指标,R2越接近1说明拟合效果越好。由表 4可知,3种模型的拟合度均较高(R2>0.99),表明选用的生长模型都能很好地描述不同饲养方式福清山羊羯羊的体重生长发育趋势与规律,其中Von Bertalanffy模型的拟合度最高,该模型A组、B组和C组福清山羊羯羊的拟合度分别高达0.999、0.999和0.998(表 4)。通过进一步利用3种数学模型计算各试验组不同时期的体重理论值(表 5),Von Bertalanffy模型计算的理论值更接近实际值,也证实该模型的拟合效果最优。
表 4 不同饲养方式下3种模型拟合参数Table 4. Fitting parameters of three models for growth patterns of Fuqing goats raised under different husbandry methods饲养方式 生长模型 A B K R2 拐点月龄 拐点体重/kg A组 Von Bertalanffy 33.583 0.533 0.145 0.999 3.24 9.95 Gompertz 31.711 2.079 0.180 0.998 4.06 11.67 Logistic 28.684 4.757 0.287 0.996 5.43 14.34 B组 Von Bertalanffy 39.960 0.542 0.116 0.999 4.19 11.84 Gompertz 36.476 2.136 0.153 0.999 4.96 13.42 Logistic 31.491 5.059 0.263 0.997 6.16 15.75 C组 Von Bertalanffy 31.564 0.505 0.141 0.998 2.95 9.35 Gompertz 29.894 1.940 0.174 0.998 3.81 11.00 Logistic 27.130 4.212 0.274 0.996 5.25 13.57 表 5 不同饲养方式下3种生长曲线模型体重估计值与实际值Table 5. Observed and model-estimated body weights of Fuqing goats raised under different husbandry methods饲养方式 生长模型 体重/kg 平均日增重/(g·d-1) 相对生长率/% 3月龄 6月龄 9月龄 12月龄 3~6月龄 6~9月龄 9~12月龄 3~6月龄 6~9月龄 9~12月龄 A组 实际值 9.32 15.98 20.67 25.09 74.0 52.1 49.1 52.6 25.6 19.3 Von Bertalanffy 9.44 15.74 21.02 25.01 70.0 58.7 44.3 50.0 28.7 17.3 Gompertz 9.44 15.65 21.01 24.95 69.0 59.6 43.8 49.5 29.2 17.1 Logistic 9.53 15.50 21.10 24.90 72.2 56.9 56.4 51.7 27.8 21.6 B组 实际值 9.33 15.83 20.95 26.03 68.1 62.7 52.4 49.2 30.7 20.1 Von Bertalanffy 9.40 15.53 21.17 25.89 67.6 63.8 51.9 48.6 31.2 19.8 Gompertz 9.46 15.54 21.28 25.95 67.2 46.8 44.7 48.9 24.1 18.6 Logistic 9.55 15.41 21.36 25.91 63.4 53 40.1 46.4 27.2 16.6 C组 实际值 9.34 15.39 19.60 23.62 62.7 53.7 39.8 45.9 27.6 16.5 Von Bertalanffy 9.46 15.17 19.94 23.55 74.0 52.1 49.1 52.6 25.6 19.3 Gompertz 9.46 15.10 19.93 23.51 70.0 58.7 44.3 50.0 28.7 17.3 Logistic 9.51 14.96 19.98 23.44 69.0 59.6 43.8 49.5 29.2 17.1 利用Von Bertalanffy模型预测A组、B组和C组的成熟体重分别为33.583、39.96和31.564 kg,拐点月龄分别为3.24、4.19和2.95,拐点体重分别为9.95、11.84和9.35 kg。同一生长模型拟合不同饲养方式的成熟体重、拐点月龄和拐点体重均呈现相同规律,为C组<A组<B组(表 3)。表明采用B组的饲养方式对福清山羊羯羊的生长发育具有促进作用,主要通过生长拐点月龄的延迟,以及拐点体重和成熟体重的增加实现。
3. 讨论
3.1 不同饲养方式对福清山羊生长发育的影响
羊只的不同饲养方式育肥效果存在差异的主要原因是摄取的营养物质不同[9]。苏铁等[4]、温琦等[10]研究表明,对放牧羔羊进行补饲可提高育肥速度。李文杨等[7]研究了放牧福清山羊生长发育情况,表明周岁内(2~12月龄)福清山羊的平均日增重为51.4 g·d-1,低于本研究的3个试验组(52.9~61.9 g·d-1),精料补饲水平的差异是造成福清山羊日增重差异的关键原因。另外,虽然本研究的试验日粮参考NRC标准,设计日增重为100 g·d-1的营养摄入水平,但3个试验组的日增重远低于预期,表明遗传背景也是影响肉羊生长发育的重要制约因素。
在日粮中补饲相同营养水平的精料,圈养福清山羊羯羊的增重效果优于放牧组,与谢喜平等[3]、武原红等[9]研究结果一致,这一方面是由于放牧条件的粗饲料来源不确定和不稳定造成的[3],另一方面不同饲养方式羊只的运动和防寒耗能不同也影响了增重效果[9]。张明等[11]研究表明,在相同日粮水平圈养下,降低饲养密度能提高小尾寒羊的生产性能;王磊等[12]也证实了圈养饲养面积越大肉羊增质量效果越好。饲喂相同日粮的前提下,B组6~12月龄的日增重高于A组,初步表明圈养福清山羊活动面积的增加会促进其生长。
3.2 不同饲养方式福清山羊生长模型拟合分析
Von Bertalanffy、Gompertz 和Logistic 3种非线性生长模型是拟合畜禽生长发育的常用的数学模型,3种模型对各试验组的拟合度均在0.99以上,拟合效果较理想,Von Bertalanffy模型的拟合效果最优。羊的生长发育遵循一定的规律,但又受到品种、生产目的、饲养管理方式等多种因素影响,不同品种间最优非线性模型也存在差异[13]。大量的研究表明Gompertz和Von Bertalanffy 模型更适合羊的生长发育拟合分析[7, 13-15],可较准确地描述大部分羊品种的生长发育规律。李文杨等[7]研究了放牧福清山羊的生长发育规律,表明Von Bertalanffy 模型对福清山羊公、母羊的拟合度高于其他模型,拟合效果最优,与本研究结果一致。
畜禽体重生长曲线是改进其饲养管理方式以及实现商品畜禽适时出栏的主要依据。采用拟合效果最优的Von Bertalanffy模型预测各试验组的成熟体重、瞬时相对生长率、拐点体重和拐点月龄均存在差异,表明饲养方式是福清山羊商品羊育肥的主要制约因素之一。B组育肥效果最优的主要原因是其生长拐点的延迟,前期快速增长时间长,而生长拐点后适当降低精料水平可减少饲料成本的支出。在饲喂日粮营养水平相似的条件下,福清山羊商品羊6月龄前采用限制活动面积进行圈养育肥,6月龄后扩大活动面积圈养育肥,体重达25 kg左右上市,可获得较理想的育肥效果和经济效益。
-
表 1 供试橄榄种质资源
Table 1 Germplasm resources of C. album used in this experiment
编号
Number名称
Germplasm
name来源地
Source经纬度
Longitude and
latitude简写名
Abbreviated
name1 葡萄 福建福安 119°38′E, 27°08′N PT 2 大目埕橄榄王 福建闽侯 118°59′E, 26°12′N DMC 3 七粒尺 福建闽侯 119°00′E, 26°10′N QLC 4 灵峰 福建闽侯 118°97′E, 26°21′N LF 5 福榄1号 福建闽侯 118°97′E, 26°21′N FL1 6 清榄1号 福建闽清 118°54′E, 26°13′N QL1 7 闽清2号 福建闽清 118°89′E, 26°24′N MQ2 8 牛榄2号 广西浦北 109°33′E, 22°19′N NL2 9 合江三白圆 四川合江 105°53′E, 28°46′N HJSBY 10 平阳3号 浙江平阳 120°30′E, 27°37′N PY3 11 瑞安2号 浙江瑞安 120°35′E, 27°53′N RA2 12 瑞安3号 浙江瑞安 120°35′E, 27°53′N RA3 13 棱尖 广东饶平 116°83′E, 23°99′N LJ 14 乌榄 广东广州 113°22′E, 23°09′N WL 表 2 测序数据统计
Table 2 Summary of sequencing data
样本名称
Sample ID全部片段数量
Number of total readsGC含量百分比
GC percentage/%Q30百分比
Q30 percentage/%DMC 749 838 40.43 93.28 FL1 894 002 41.03 93.43 HJSBY 7 541 767 37.41 93.43 LF 559 479 39.21 93.54 LJ 1 012 589 38.36 93.61 MQ2 1 139 514 40.38 93.51 NL2 1 071 660 38.80 93.62 PT 876 458 41.65 93.48 PY3 964 858 39.20 93.32 QL1 1 996 699 40.74 93.44 QLC 813 057 41.57 93.61 RA2 1 128 676 39.18 93.76 RA3 873 826 42.05 93.67 WL 901 408 37.88 93.29 表 3 不同样本中SNP位点数量
Table 3 Number of SNP loci in different samples
样本名称
Sample IDSNP数量
SNP number完整度
Hetloci ratio/%杂合度
Integrity ratio/%DMC 227 472 13.96 62.94 FL1 239 971 14.73 66.40 HJSBY 297 219 21.86 82.24 LF 209 271 12.95 57.90 LJ 255 598 17.94 70.72 MQ2 249 658 14.61 69.08 NL2 256 355 17.77 70.93 PT 231 235 14.92 63.98 PY3 239 403 15.00 66.24 QL1 290 076 30.22 80.26 QLC 233 083 13.59 64.49 RA2 254 930 16.64 70.54 RA3 236 723 15.11 65.50 WL 165 152 11.72 45.69 -
[1] XIANG Z B, WU X L, LIU X Y. Chemical composition and antioxidant activity of petroleum ether extract of Canarium album [J]. Pharmaceutical Chemistry Journal, 2017, 51(7): 606−611. DOI: 10.1007/s11094-017-1661-9
[2] DUAN W J, TAN S Y, CHEN J, et al. Isolation of anti-HIV components from Canarium album fruits by high-speed counter-current chromatography [J]. Analytical Letters, 2013, 46(7): 1057−1068. DOI: 10.1080/00032719.2012.749486
[3] JIA Y L, ZHENG J, YU F, et al. Anti-tyrosinase kinetics and antibacterial process of caffeic acid N-nonyl ester in Chinese olive (Canarium album) postharvest [J]. International Journal of Biological Macromolecules, 2016, 91: 486−495. DOI: 10.1016/j.ijbiomac.2016.05.098
[4] LIU Q P, ZHOU M L, ZHENG M, et al. Canarium album extract restrains lipid excessive accumulation in hepatocarcinoma cells [J]. International Journal of Clinical and Experimental Medicine, 2016, 9(9): 17509−17518.
[5] 赖瑞联, 陈瑾, 韦晓霞, 等. 中国橄榄研究40年 [J]. 热带作物学报, 2020, 41(10):2045−2054. DOI: 10.3969/j.issn.1000-2561.2020.10.011 LAI R L, CHEN J, WEI X X, et al. Research of Chinese olive in the past 40 years [J]. Chinese Journal of Tropical Crops, 2020, 41(10): 2045−2054. (in Chinese) DOI: 10.3969/j.issn.1000-2561.2020.10.011
[6] 杨培奎. 粤东地区橄榄种质资源遗传多样性ISSR分析及核心种质初步构建[D]. 汕头: 汕头大学, 2010. YANG P K. Analysis of genetic diversity of Canarium album L. germplasm resources from East Guangdong Province using ISSR and construction of core collection[D]. Shantou: Shantou University, 2010. (in Chinese)
[7] 刘天亮. 福建省橄榄(Canavium album Raeusch)种质资源的ISSR分析[D]. 福州: 福建农林大学, 2010. LIU T L. ISSR analysis of germplasm resources of Fujian Canavium album Raeusch[D]. Fuzhou: Fujian Agriculture and Forestry University, 2010. (in Chinese)
[8] 赖瑞联, 陈瑾, 冯新, 等. 橄榄ISSR和RAPD遗传多样性分析和核心种质构建 [J]. 热带亚热带植物学报, 2022, 30(1):41−53. DOI: 10.11926/jtsb.4437 LAI R L, CHEN J, FENG X, et al. ISSR and RAPD genetic diversity analysis and core germplasms construction of Canarium album [J]. Journal of Tropical and Subtropical Botany, 2022, 30(1): 41−53. (in Chinese) DOI: 10.11926/jtsb.4437
[9] 聂珍素. 福建橄榄(Canarium album L. )资源的RAPD分析[D]. 福州: 福建农林大学, 2005. NIE Z S. RAPD analysis of album(Canarium album) genetic resources in Fujian Province[D]. Fuzhou: Fujian Agriculture and Forestry University, 2005. (in Chinese)
[10] MEI Z L, ZHANG X Q, LIU X Y, et al. Genetic analysis of Canarium album in different areas of China by improved RAPD and ISSR [J]. Comptes Rendus Biologies, 2017, 340(11/12): 558−564.
[11] MEI Z Q. Improved RAPD analysis of Canarium album (Lour. ) raeusch from Sichuan Province along Yangtze River in China [J]. Annual Research & Review in Biology, 2014, 4: 51−60.
[12] CHENG J L, YIN Z C, MEI Z Q, et al. Development and significance of SCAR marker QG12-5 for Canarium album (Lour. ) Raeusch by molecular cloning from improved RAPD amplification [J]. Genetics and Molecular Research, 2016, 15(3): gmr8347.
[13] 张小红, 赵依杰, 林航. 基于SRAP技术的甜橄榄种质资源遗传多样性分析 [J]. 中国南方果树, 2017, 46(6):53−56. ZHANG X H, ZHAO Y J, LIN H. Genetic diversity analysis of sweet olive germplasm resources based on SRAP technology [J]. South China Fruits, 2017, 46(6): 53−56. (in Chinese)
[14] 张平湖, 刘冠明. 橄榄SRAP-PCR体系的建立和优化 [J]. 中国农学通报, 2010, 26(15):86−88. ZHANG P H, LIU G M. Establishment and optimization of SRAP-PCR system in Canarium album reausch [J]. Chinese Agricultural Science Bulletin, 2010, 26(15): 86−88. (in Chinese)
[15] 王燕平, 陈勤, 周志钦, 等. 橄榄基因组AFLP扩增体系的优化 [J]. 果树学报, 2012, 29(3):505−511. WANG Y P, CHEN Q, ZHOU Z Q, et al. Optimization of AFLP reaction system in Canarium album [J]. Journal of Fruit Science, 2012, 29(3): 505−511. (in Chinese)
[16] 苏睿, 林峻, 陈鲤群, 等. 高通量自动化SNP检测技术研究进展 [J]. 中国细胞生物学学报, 2019, 41(7):1412−1422. DOI: 10.11844/cjcb.2019.07.0022 SU R, LIN J, CHEN L Q, et al. Research progress on high-throughput automated SNP detection technology [J]. Chinese Journal of Cell Biology, 2019, 41(7): 1412−1422. (in Chinese) DOI: 10.11844/cjcb.2019.07.0022
[17] ZHU Z S, SUN B M, LEI J J. Specific-locus amplified fragment sequencing (SLAF-seq) as high-throughput SNP genotyping methods [J]. Methods in Molecular Biology, 2021, 2264: 75−87.
[18] 赖瑞联, 徐洋, 赖恭梯, 等. 山枇杷基因组DNA的提取及自然群体内ISSR遗传多样性分析 [J]. 森林与环境学报, 2015, 35(1):53−59. LAI R L, XU Y, LAI G T, et al. Extraction of genomic DNA and analysis of genetic diversity of the natural population by ISSR in Garcinia multiflora [J]. Journal of Forest and Environment, 2015, 35(1): 53−59. (in Chinese)
[19] SUN X W, LIU D Y, ZHANG X F, et al. SLAF-seq: An efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing [J]. PLoS One, 2013, 8(3): e58700. DOI: 10.1371/journal.pone.0058700
[20] XU Q, CHEN L L, RUAN X A, et al. The draft genome of sweet orange (Citrus sinensis) [J]. Nature Genetics, 2013, 45: 59−66. DOI: 10.1038/ng.2472
[21] KOZICH J J, WESTCOTT S L, BAXTER N T, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform [J]. Applied and Environmental Microbiology, 2013, 79(17): 5112−5120. DOI: 10.1128/AEM.01043-13
[22] TAMURA K, PETERSON D, PETERSON N, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods [J]. Molecular Biology and Evolution, 2011, 28(10): 2731−2739. DOI: 10.1093/molbev/msr121
[23] ALEXANDER D H, NOVEMBRE J, LANGE K. Fast model-based estimation of ancestry in unrelated individuals [J]. Genome Research, 2009, 19(9): 1655−1664. DOI: 10.1101/gr.094052.109
[24] 赵亚琴, 樊丛照, 张际昭, 等. 基于简化基因组技术的啤酒花栽培种和野生种SNP位点开发及遗传结构分析 [J]. 中草药, 2021, 52(20):6365−6372. DOI: 10.7501/j.issn.0253-2670.2021.20.027 ZHAO Y Q, FAN C Z, ZHANG J Z, et al. SNP loci development and genetic structure of cultivated and wild individuals of Humulus lupulus using SLAF-seq [J]. Chinese Traditional and Herbal Drugs, 2021, 52(20): 6365−6372. (in Chinese) DOI: 10.7501/j.issn.0253-2670.2021.20.027
[25] 田倩, 刘双委, 钮世辉, 等. 基于SLAF-seq技术的白皮松SNP分子标记开发 [J]. 北京林业大学学报, 2021, 43(8):1−8. DOI: 10.12171/j.1000-1522.20200211 TIAN Q, LIU S W, NIU S H, et al. Development of SNP molecular markers of Pinus bungeana based on SLAF-seq technology [J]. Journal of Beijing Forestry University, 2021, 43(8): 1−8. (in Chinese) DOI: 10.12171/j.1000-1522.20200211
[26] CHEN Z Y, HE Y C, IQBAL Y, et al. Investigation of genetic relationships within three Miscanthus species using SNP markers identified with SLAF-seq [J]. BMC Genomics, 2022, 23(1): 43. DOI: 10.1186/s12864-021-08277-8
[27] WEI Q Z, WANG W H, HU T H, et al. Construction of a SNP-based genetic map using SLAF-seq and QTL analysis of morphological traits in eggplant [J]. Frontiers in Genetics, 2020, 11: 178. DOI: 10.3389/fgene.2020.00178
[28] WANG R J, GAO X F, YANG J, et al. Genome-wide association study to identify favorable SNP allelic variations and candidate genes that control the timing of spring bud flush of tea (Camellia sinensis) using SLAF-seq [J]. Journal of Agricultural and Food Chemistry, 2019, 67(37): 10380−10391. DOI: 10.1021/acs.jafc.9b03330
[29] 樊晓静, 于文涛, 蔡春平, 等. 利用SNP标记构建茶树品种资源分子身份证 [J]. 中国农业科学, 2021, 54(8):1751−1772. DOI: 10.3864/j.issn.0578-1752.2021.08.014 FAN X J, YU W T, CAI C P, et al. Construction of molecular ID for tea cultivars by using of single-nucleotide polymorphism(SNP) markers [J]. Scientia Agricultura Sinica, 2021, 54(8): 1751−1772. (in Chinese) DOI: 10.3864/j.issn.0578-1752.2021.08.014
[30] 魏中艳, 李慧慧, 李骏, 等. 应用SNP精准鉴定大豆种质及构建可扫描身份证 [J]. 作物学报, 2018, 44(3):315−323. DOI: 10.3724/SP.J.1006.2018.00315 WEI Z Y, LI H H, LI J, et al. Accurate identification of varieties by nucleotide polymorphisms and establishment of scannable variety IDs for soybean germplasm [J]. Acta Agronomica Sinica, 2018, 44(3): 315−323. (in Chinese) DOI: 10.3724/SP.J.1006.2018.00315
-
期刊类型引用(1)
1. 王哲,邓乐平,龙永彬,谢威,曾明,欧阳曦,郭文冰,李义良. 基于SLAF-seq技术的湿加松遗传多样性和遗传结构分析. 林业与环境科学. 2025(01): 1-8 .  百度学术
百度学术
其他类型引用(0)




 下载:
下载: