Research Progress on Effects of Plant Hormones and Other Regulatory Factors on Rice Root Growth and Development
-
摘要:
水稻(Oryza sativa L.)作为全世界重要的粮食作物,提高其产量是保障国家粮食安全的重要策略。根系作为水稻最重要的器官之一,在水稻的生长发育、信号感知、激素合成、水分和养分吸收以及产量形成中起重要作用。然而,水稻根系的形成及生长发育也受到多种因素的影响。其中,植物激素在调控水稻根系生长发育过程中发挥着重要作用,使植物能够根据环境需求调节其发育、生长和生理状态。本文综述了影响水稻根系生长发育的非生物因素和生物因素;概述了与根系生长相关的基因及调控网络,重点阐述了植物激素在水稻根系形成及生长发育中的作用机制;总结了几种激素之间相互协同对根系的生长调控,并对其他影响根系的小分子物质进行讨论,旨在为水稻根系研究及提高水稻产量提供理论依据,也为今后更加精细化研究激素信号通路之间复杂的串扰以及和其他信号之间的作用机制提供新方向。
Abstract:Being one of the major food sources in the world, rice (Oryza sativa L.) has been studied to promote its growth and increase its production for the security of human livelihood. Since the root system is crucial for the growth, signal perception, hormone synthesis, water and nutrient uptakes, and production of a rice plant, the formation, development, and functions of the system need to be well understood. However, since the affecting factors are numerous, the study could be challenging. For instance, plant hormones are of special interest regarding their roles in regulating root growth and development under ever-changing environmental conditions. This article comprehensively reviewed subjects involving abiotic and biotic factors, related genes and regulatory networks, regulating plant hormones, and synergisms among hormones, metabolic trace minerals, and low-molecular substances. By emphasizing the studies on the plant hormones, it is hoped that research to further rice yield improvement and in-depth understanding of the basic biological mechanisms, complex interactions/interferences among biochemicals, and functional signaling pathways associated with rice root system development and functions will be realized.
-
Keywords:
- rice /
- root structure /
- plant hormones /
- regulatory network /
- growth and development
-
0. 引言
【研究意义】木薯(Manihot esculenta Crantz)为大戟科木薯属直立灌木状多年生热带作物,起源于拉丁美洲[1],被广泛种植于热带和亚热带地区,是全球第六大粮食作物[2],为全球10亿多人提供了主要的食物来源[3]。木薯在种植过程中,面临着多种逆境胁迫,其中木薯花叶病(Cassava mosaic disease, CMD)是木薯最严重的病毒性疾病之一,可导致木薯高达95%的产量损失,是木薯生产的主要制约因素[4]。鉴于此,运用现代生物技术来发掘与逆境胁迫相关的基因,增强木薯的抗逆性和抗病性,显得尤其重要。【前人研究进展】研究人员通过全基因组关联研究(Genome-wide association studies, GWAS)和双亲连锁图谱发现CMD的抗性主要是由12号染色体上的一个主要基因座赋予的,该基因被称为CMD2基因座[5,6]。同时,Rabbi等研究发现CMD2基因座中的两个候选基因MePOD10(Manes.12G076200)和MePOD58(Manes.12G076300)与CMD的抗性紧密相关[7]。过氧化物酶是广泛存在于各种动物、植物和微生物体内的一类氧化酶[8]。其按照来源可分为3类,分别是Class Ⅰ、Class Ⅱ和Class Ⅲ[9]。第一类POD(Class Ⅰ POD),作为广泛存在于植物、真菌以及原核生物中的一种过氧化物酶,其主要功能是清除体内过多的过氧化氢,并在此过程中调节细胞的氧化还原稳态[10]。第二类POD(Class Ⅱ POD)主要存在于真菌中,并在木质素的降解过程中发挥着至关重要的作用[11]。第三类POD(Class Ⅲ POD)是植物中特有的分泌型过氧化物酶,在抵抗逆境胁迫的过程中发挥着重要作用[12]。在植物的生长及发育过程中,Class Ⅲ POD涉及了很多的生理生化反应,如参与生长素的分解代谢、细胞壁的合成代谢[13]、活性氮的代谢[14]、防御病原体[15]、木质素化和木栓形成[16]、活性氧(Reactive oxygen species, ROS)清除、伤口愈合和植物抗毒素合成等对生物和非生物胁迫的防御反应[17]。【本研究切入点】前人通过GWAS在CMD2基因座找到的MePOD10和MePOD58尚未在木薯中探究其功能,且有关木薯POD基因克隆和功能分析的研究相对较少。【拟解决的关键问题】本研究拟克隆MePOD10基因,利用生物信息学工具对其编码蛋白序列进行分析,并构建原核表达载体确定其蛋白表达条件,为深入探究MePOD10蛋白的功能奠定理论基础。
1. 材料与方法
1.1 试验材料
选用木薯品种SC124作为试验材料,其种植于海南大学儋州校区种植实验苗圃基地(19°50'N, 109°49'E),原核表达载体pET28a由实验室自行保存。植物总RNA提取试剂盒、异丙基-β-D-硫代吡喃半乳糖苷(IPTG)、DNA回收试剂盒购自天根生化科技有限公司、His标签蛋白纯化试剂盒购自上海碧云天生物技术股份有限公司、POD酶活测定试剂盒购自南京建成生物工程研究所、愈创木酚购自上海麦克林生化科技股份有限公司等。
1.2 木薯总RNA的提取和反转录
从SC124健康植株上剪取适量新鲜叶片,将其剪碎置于研钵内,加入少许液氮研磨成细腻粉状,经RNA提取试剂盒提取木薯总RNA后,再反转为cDNA,−80 ℃保存备用。
1.3 基因的克隆
在Phytozome网站(https://phytozome.jgi.doe.gov)查询MePOD10基因(Manes.12G076300)的CDS序列,用Primer 5.0软件设计特异性引物(表1),用cDNA作为模板扩增MePOD10基因CDS序列。经电泳跑胶验证PCR产物有目的条带后,切胶回收目的条带并将其构建于pEASY-Blunt3载体上,转化后挑选边缘清晰、大小适中且明亮的单菌落,摇菌并提取质粒送测序,将测序正确的单克隆菌液于−40 ℃保存备用。
表 1 供试引物序列Table 1. Primers applied引物名称
Primer name引物序列
Primer sequence引物用途
Primer usageMePOD10-F
MePOD10-R5'-GGAATTCATGGGAGGTTTTGGTTCTTT-3'
5'-GGATCCTCAACTATTGATCACTGCAC-3'基因克隆
Gene cloningMePOD10-pET28a-F
MePOD10-pET28a-R5'-ATGGGTCGCGGATCCATGGGAGGTTTTGGTTCTTT-3'
5'-GTCGACGGAGCTCGAATTCCAGGTCCTCCTCTGAGA
TCAGCTTCTGCTCCTCACTATTGATCACTGCAC-3'原核表达
Prokaryotic expression1.4 生物信息学分析
MePOD10基因编码的蛋白理化特性由ExPASy(http://web.expasy.org/protparam/)分析,MePOD10蛋白保守结构域利用NCBI(https://www.ncbi.nlm.nih.gov/)的CDD数据库分析。MePOD10蛋白的二级和三级结构利用Prabi(https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html)和SWISS-MODEL(https://swissmodel.expasy.org/interactive)预测。通过NCBI数据库BLAST搜索MePOD10基因同源序列,并确定其开放阅读框(Open reading frame, ORF)。由MEGA7.0翻译MePOD10基因CDS序列,与其他物种进行同源比对分析后构建系统发育进化树。
1.5 原核表达载体的构建
以MePOD10-Blunt3质粒为模板,扩增MePOD10-pET28a片段,将扩增好的片段以同源重组方法构建于pET28a载体上,转化后挑选边缘清晰、大小适中且明亮的单菌落,摇菌并提取质粒送测序,将测序正确的单克隆菌液−40 ℃保存备用。
1.6 重组蛋白的诱导表达
将构建成功的MePOD10-pET28a质粒转入BL21(DE3)感受态细胞中,选取PCR检测无误的单克隆作为表达菌,将表达菌接种于LB培养基中,利用紫外分光光度计将表达菌液初始D600调至0.2~0.3,置于37 ℃摇床,表达菌液摇至D600为0.6~0.8,加入IPTG至终浓度为1 mmol·L−1并置于37 ℃摇床,200 r·min−1条件下诱导。取诱导6 h的重组蛋白为样品,经离心后取上清和沉淀加入上样缓冲液后充分涡旋,SDS-PAGE检测。
1.7 重组蛋白的Western blotting验证
经SDS-PAGE电泳后,进行转膜,转完的膜经1×TBS 漂洗后置于15 mL的脱脂牛奶中,室温下摇床轻晃1 h。每隔10 min用适量的1×TBS洗膜3次,加入Myc抗体室温孵育2 h,每隔10 min用适量的1×TBS洗膜3次,加入二抗室温孵育1 h,最后用A、B显影液进行显影成像。
1.8 重组蛋白的纯化和酶活测定
选用上述诱导条件对MePOD10重组蛋白进行大量诱导,将诱导后的菌液离心,用磷酸缓冲溶液重悬菌体,功率25%、冰上超声破碎10 min后离心得到的上清用试剂盒纯化。用POD酶活试剂盒测定MePOD10蛋白酶活性,pET28a空载蛋白作对照,重复3次。以不同浓度愈创木酚(0、10、20、30、40、50 mmol·L−1)为底物设置5组试验,以磷酸缓冲溶液作对照调零,在反应温度37 ℃、pH5.8条件下,取5 mL反应混合液(磷酸缓冲溶液3 mL、不同浓度的愈创木酚1 mL和1 mL 2%过氧化氢,摇匀),每组各加入1 mL纯化后的MePOD10蛋白,立即摇匀,并迅速倒入比色皿中,于470 nm波长下比色,1 min后记录各组吸光度值∆A,重复3次。按下式计算酶反应初始速度V0:
V0=ΔA⋅Vtε⋅Δt⋅Vs⋅c ∆A为反应初速度阶段内吸光度的变化;Vt为提取酶液总体积(mL);ε为愈创木酚的摩尔吸光系数(mol·L−1·cm−1);∆t为反应时间;Vs为测定时取用蛋白体积(mL);c为蛋白浓度(mg·mL−1)。使用GraphPad Prism 8.0拟合酶动力学曲线并进行单因素方差分析。
2. 结果与分析
2.1 MePOD10基因克隆及鉴定
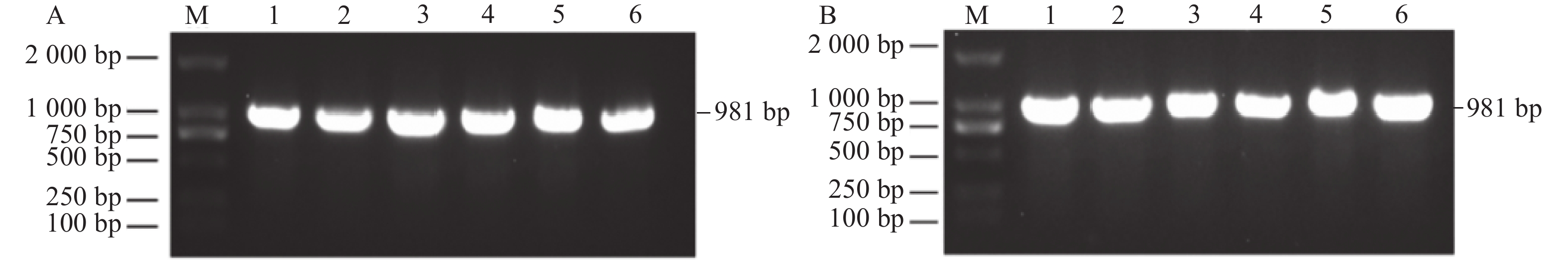
提取SC124木薯新鲜叶片总RNA并反转录成cDNA,以cDNA为模板进行PCR扩增,得到目的基因片段(图1A)。回收的目的片段构建于pEASY-Blunt3载体上,转入DH5α菌株中,菌落PCR选取阳性克隆(图1B),摇菌并提取质粒送测序。经序列比对显示,该阳性克隆长度为981 bp,编码326个氨基酸,鉴定为MePOD10基因。
2.2 生物信息学分析
2.2.1 MePOD10编码蛋白理化性质和保守结构域分析
使用在线工具Expasy对MePOD10蛋白进行预测分析,结果显示MePOD10的分子式为C
1534 H2434 N434O481S13,分子量为35069.60 Da,接近中性的理论等电点为6.59。Prabi预测结果显示,MePOD10蛋白由α-螺旋、伸展链和不规则卷曲组成蛋白二级结构。MePOD10蛋白不稳定性指数为42.22,GRAVY指数(总平均疏水性)为−0.067,表明其是一种不稳定疏水性蛋白。WoLF PSORT II预测结果显示,MePOD10基因主要定位于细胞核膜上。NCBI-CDD数据库对MePOD10蛋白保守结构域预测,结果显示在24~324 bp区间存在一个POD保守结构域。2.2.2 多序列比对与进化分析
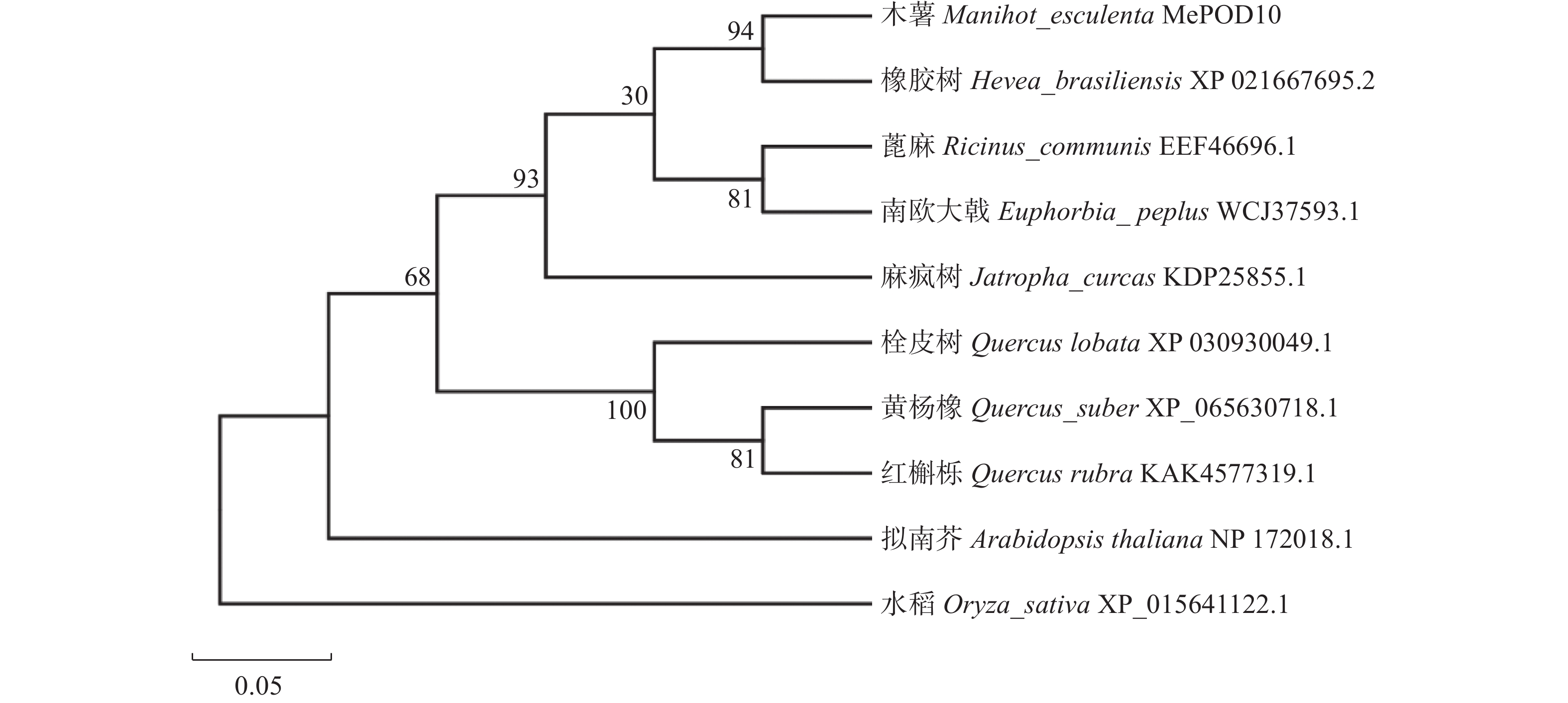
通过NCBI数据库中的BLAST工具对MePOD10的氨基酸序列进行比对分析,筛选与木薯MePOD10蛋白具有较高同源性的氨基酸序列,结果(图2)显示MePOD10与橡胶树(Hevea brasiliensis; XP_021667695.2)的POD蛋白氨基酸序列同源性最高,为93.25%。此外,与麻疯树(Jatropha curcas; KDP25855.1)和蓖麻(Ricinus communis; EEF46696.1)的同源性也较高,分别为88.04%和87.93%。系统发育进化树显示,MePOD10与橡胶树的POD蛋白亲缘关系最近(图3),表明两者在进化过程中保持了较高的序列相似性和功能保守性。
2.3 MePOD10原核表达载体的构建及鉴定
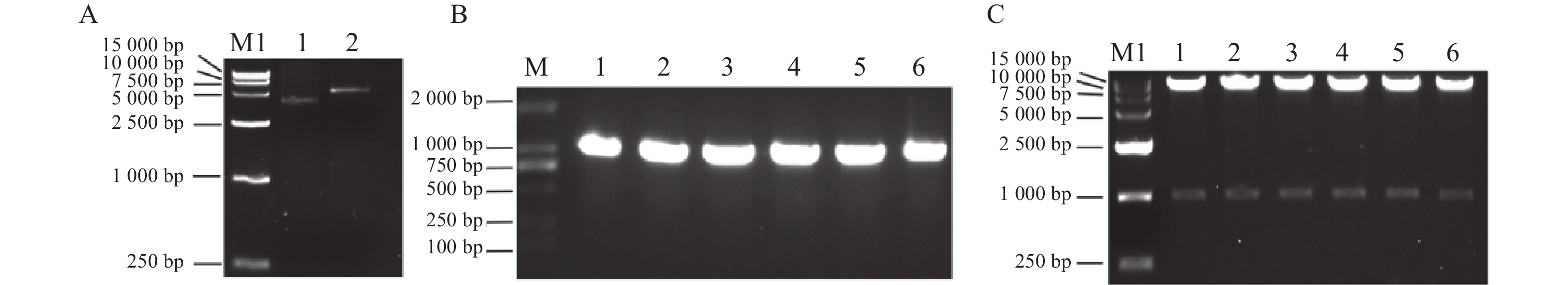
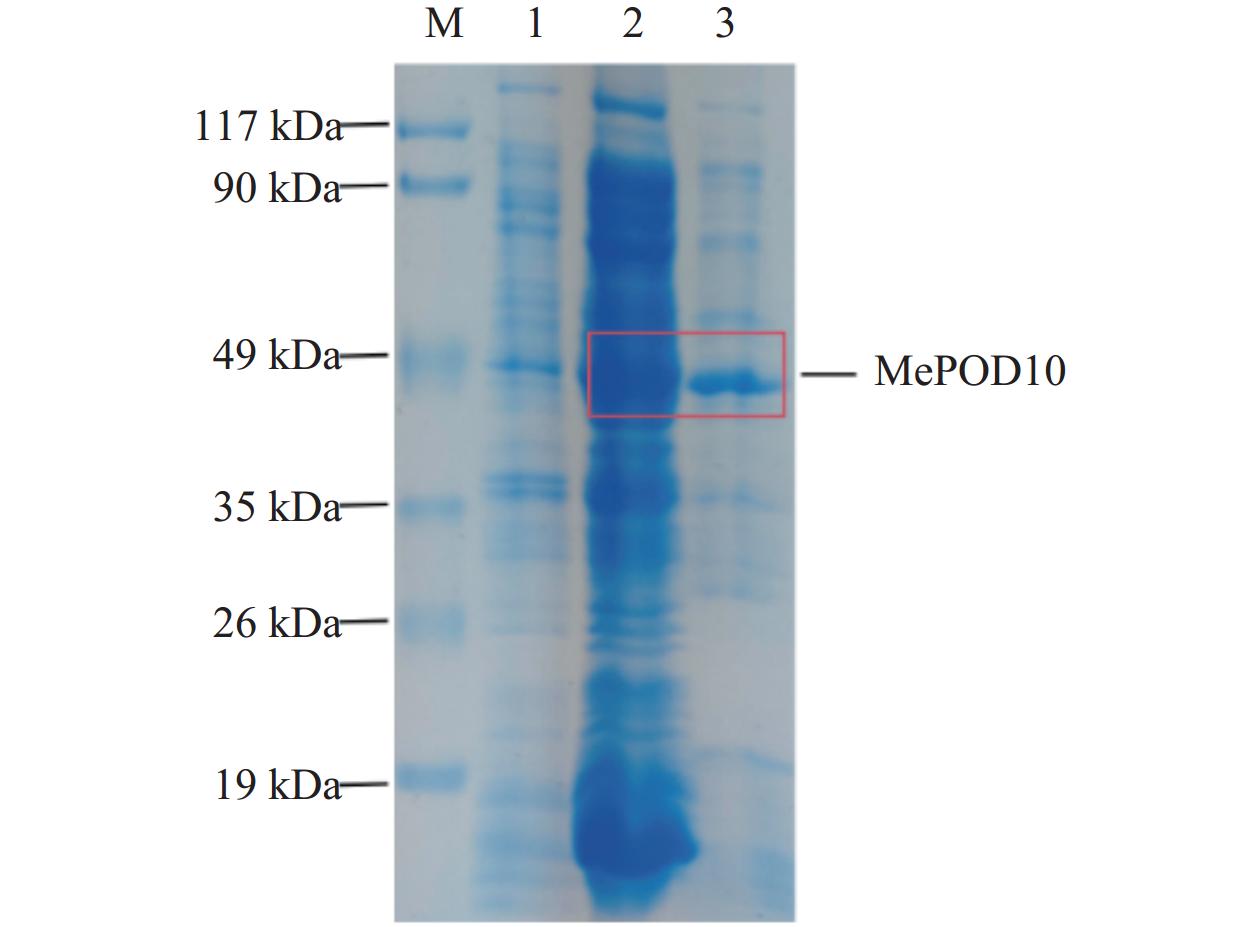
利用同源重组方法将MePOD10基因构建于EcoRⅠ和BamHⅠ双酶切好的pET28a载体(图4A)上,再转化至DH5α感受态细胞中,涂板后挑取6个单克隆进行PCR验证,得到条带大小与目的片段一致的阳性克隆(图4B),将阳性单克隆摇菌后碱裂解法提取质粒,再双酶切质粒验证(图4C)。
![]() 图 4 MePOD10-pET28a载体构建M:DL2000 DNA标记,M1: 15000 DNA标记;A:1为pET28a空载,2为酶切pET28a;B:同源重组后菌落PCR;1~6为PCR检验;C:1~6为酶切6个MePOD10-pET28载体。Figure 4. Construction of MePOD10-pET28a vectorM: DL2000 DNA marker; M1: 15,000 DNA marker; A: 1: empty vector of pET28a; 2: enzyme-digested pET28a; B: PCR of homologous recombinant colonies; 1-6: PCR tests; C: 6 digested MePOD10-pET28a vectors.
图 4 MePOD10-pET28a载体构建M:DL2000 DNA标记,M1: 15000 DNA标记;A:1为pET28a空载,2为酶切pET28a;B:同源重组后菌落PCR;1~6为PCR检验;C:1~6为酶切6个MePOD10-pET28载体。Figure 4. Construction of MePOD10-pET28a vectorM: DL2000 DNA marker; M1: 15,000 DNA marker; A: 1: empty vector of pET28a; 2: enzyme-digested pET28a; B: PCR of homologous recombinant colonies; 1-6: PCR tests; C: 6 digested MePOD10-pET28a vectors.2.4 MePOD10重组蛋白的原核表达
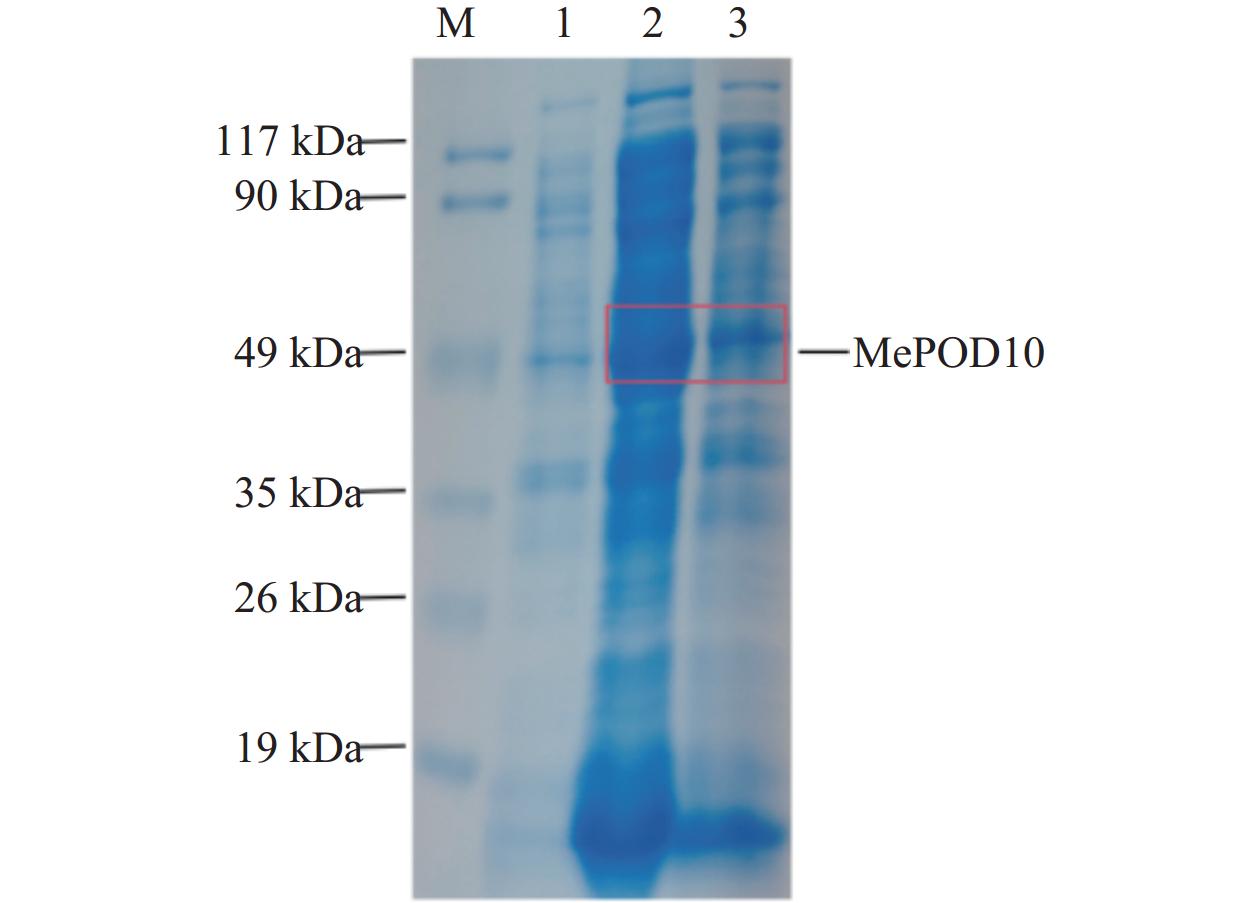
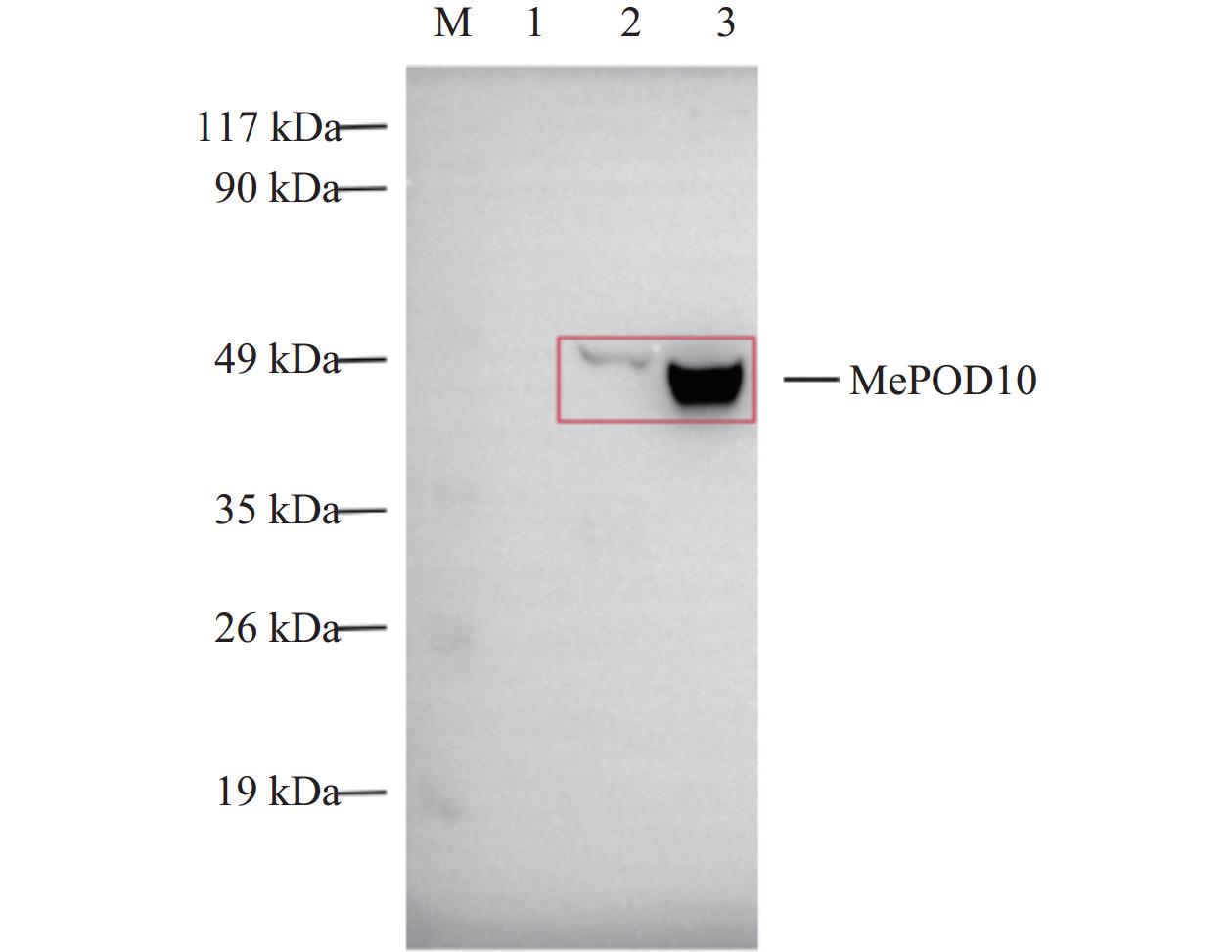
将验证正确的重组质粒转入BL21(DE3)感受态细胞中,在诱导温度为37 ℃,D600值为0.6~0.8条件下,加入IPTG至终浓度为1 mmol·L−1,并在200 r·min−1条件下培养。诱导蛋白表达6 h后,进行SDS-PAGE凝胶电泳检测。结果发现,在49 kDa之间有特异性条带,与预期的MePOD10重组蛋白大小一致(图5)。
2.5 MePOD10重组蛋白的Western blotting验证
为进一步明确MePOD10重组蛋白表达的准确性,本研究利用Myc抗体,对收集的重组蛋白上清和沉淀进行Western blotting检测。结果表明:重组蛋白在49 kDa处出现特异性条带,说明重组蛋白表达成功(图6)。
2.6 MePOD10重组蛋白的纯化及酶活性测定
根据上述诱导条件对MePOD10重组蛋白大量诱导及纯化,获得纯化的蛋白(图7),大小与MePOD10重组蛋白大小一致,约49 kDa。由MePOD10重组蛋白的酶活性检测结果(图8A)可知,MePOD10蛋白酶活性显著高于对照蛋白(P<0.0001),说明MePOD10重组蛋白具有催化活性。以愈创木酚为底物的MePOD10蛋白酶动力学特性结果(图8B)表明,随着愈创木酚浓度增加,蛋白催化活性迅速增加,后趋于平稳,说明MePOD10蛋白具有POD酶活性。MePOD10蛋白Km值为3.113 mmol·L−1,表明MePOD10蛋白对愈创木酚亲和力大,酶促反应容易进行。
![]() 图 8 MePOD10蛋白酶活性和动力学分析A:对照蛋白(pET28a)和MePOD10蛋白酶活测定;B:以愈创木酚为底物时,MePOD10蛋白酶学动力曲线。****表示差异显著(P<0.0001)。Figure 8. Enzymatic activity and kinetic analysis of MePOD10A: enzymatic activities of pET28a (control) and MePOD10; B: kinetics of MePOD10 protein observed with guaiacol as substrate. ****: significant difference at P<0.0001.
图 8 MePOD10蛋白酶活性和动力学分析A:对照蛋白(pET28a)和MePOD10蛋白酶活测定;B:以愈创木酚为底物时,MePOD10蛋白酶学动力曲线。****表示差异显著(P<0.0001)。Figure 8. Enzymatic activity and kinetic analysis of MePOD10A: enzymatic activities of pET28a (control) and MePOD10; B: kinetics of MePOD10 protein observed with guaiacol as substrate. ****: significant difference at P<0.0001.3. 讨论
随着全球气候的变化,木薯在生长发育的过程中受到各种胁迫与病害的制约,其中木薯花叶病是木薯最严重的病害之一。从经济、环保、稳定性和持久性等长远目光来看,常规分子育种是应对花叶病的有效长期策略[18]。
目前的木薯分子育种研究不仅挖掘到了针对CMD的单基因显性抗性CMD2基因座,还发现了另外两种抗性来源:多基因隐性抗性基因座CMD1以及与CMD2不关联的CMD3基因座。携带CMD2抗性位点的木薯几乎都表现出对木薯花叶病的高度抗性[19],随后有相关研究在CMD2基因座中找到了两个候选基因MePOD10和MePOD58,发现其与木薯花叶病的抗性紧密相关,但其具体的抗病功能还未得到验证。为进一步研究MePOD10在木薯中抗花叶病的功能,本研究构建了原核表达载体MePOD10-pET28a。原核表达载体有多种系列,其中pET是目前使用最为广泛的载体之一[20]。BL21(DE3)也是一种高效且成本较低的表达系统[21]。本研究所选用的pET28a载体和大肠杆菌 BL21(DE3)可以正确、大量地诱导MePOD10蛋白表达,该融合蛋白大小约为49 kDa,符合预测大小。此外,本研究发现在1 mmol·L−1 IPTG、37 ℃、200 r·min−1条件下诱导6 h可以得到较多的目的蛋白,且上清和沉淀均能表达出蛋白。相关研究表明影响大肠杆菌表达的因素有时间、温度等多方面,且37 ℃为大肠杆菌诱导的适宜温度[22]。
POD是植物体内一种非常重要且普遍存在的酶类。POD在植物体内扮演着多重角色,特别是在氧化还原反应和细胞保护方面发挥着关键作用,是植物体内的主要保护酶之一[23]。POD活性随植物生长发育进程以及环境条件的改变而变化,在逆境条件下(如干旱、盐胁迫、病虫害等),植物体内会产生大量的活性氧,导致氧化胁迫加剧。此时,POD等抗氧化酶的活性会显著增强,以清除植物体内过剩的活性氧,减轻氧化损伤的程度。因此,测定POD活性的高低可以作为衡量植物抗逆性强弱的一个重要指标。POD酶活性测定方法有多种,使用广泛且操作简便的方法是愈创木酚法[24]。在酶动力学研究中,Km值是酶特征性常数,可以代表酶与底物的亲和能力,且其大小与酶的性质有关,Km越小,表明酶与底物的亲和力越高,催化作用越大[25]。本研究以不同浓度的愈创木酚为底物时测定MePOD10活性,拟合酶动力学曲线发现,随着底物浓度增加,酶促反应逐渐上升,后趋于平稳,且Km值为3.113 mmol·L−1,表明MePOD10具有POD活性且对愈创木酚的亲和力和催化作用大。本研究初步测定MePOD10蛋白酶活性并拟合酶动力学曲线计算Km值,为进一步探究其蛋白功能奠定基础。
4. 结论
本研究对MePOD10基因进行克隆、原核表达分析以及蛋白酶活性和动力学分析研究,并且对其进行了生物信息学和原核表达情况的初步分析,探索了一些目的蛋白的诱导方法和条件,具有一定的前瞻性和创新性。由于目的蛋白在上清表达量相对沉淀较少,在进一步纯化蛋白试验中目的蛋白纯度较低,因此诱导目的蛋白的更优条件仍有待深入发掘。
-
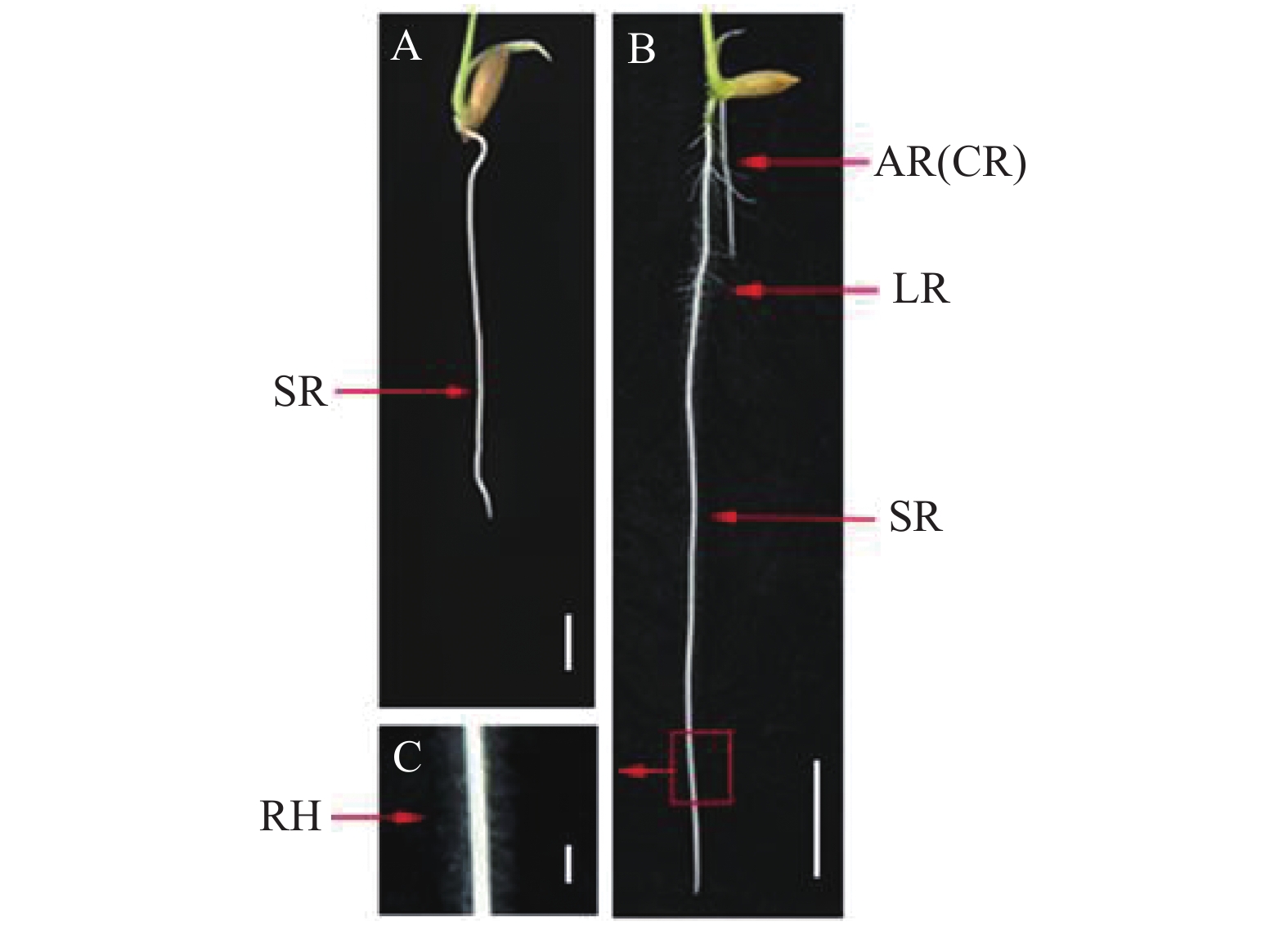
图 1 水稻根系组成[18]
A:4日龄水稻幼苗的根系, 标尺= 0.5 cm;B:7日龄水稻幼苗的根系,标尺= 1 cm;C:水稻根毛,标尺= 1 mm。SR:初生根;AR: 不定根;CR:冠根;RH:根毛。
Figure 1. Root system of a rice plant [18]
A: root of a 4-d-old rice seeding, scale bar=0.5 cm; B: root of a 7-d-old rice seeding, scale bar=1 cm; C: root hairs of a rice plant, scale bar=1 mm; SR: seminal root; AR: adventitious root; CR: crown root; RH: root hair.
表 1 水稻中已克隆的根系生长发育相关基因及其功能
Table 1 Cloned genes and functions associated with rice root growth and development
基因符号
Gene symbol基因号
Genomic Locus突变体表型
Mutant phenotype参考文献
ReferencesOsA8 Os03g0100800 OsA8的敲除突变植株根系生物量减少 [1] OsPHR3 Os02g0139000 OsPHR3突变体不定根和根毛发育减弱 [2] OsWRKY74 Os09g0334500 过表达植株在Pi或是P缺乏时,根的生物量更高 [3] OsAMT1;3 Os02g0620500 低铵条件下,敲除突变体植株总侧根数、种子根和侧根长度均降低 [4] OsGRX6 Os01g0667900 过表达OsGRX6的植株根干重下降 [5] OsPHO2 Os05g0557700 OsPHO2的敲除突变体植株根系更短 [6] GCN5 Os10g0415900 GCN5敲除突变体植株冠状根较少,根长降低 [7] OsAIP1 Os01g0125800 OsAIP1过表达和敲除植株根毛长度较短,直径增大 [8] OsCYCP4;1 Os10g0563900 过表达OsCYCP4;1植株根长度较短 [9] OsCSLD1 Os10g0578200 OsCSLD1突变体植株根毛长度偏短、局部扭曲膨胀 [10] RT Os04g0497200 突变体rt根长较短 [11] OsDRP2B Os02g0738900 突变体主根长度变短,侧根直径增加 [12] OsTSD2 Os02g0755000 突变体初生根较短、侧根更密 [13] OsPUB15 Os08g0110500 OsPUB15突变体种子根缺失 [14] OsCatB Os06g0727200 过表达CatB植株根重下降 [15] miR390 Os03g0817300 过表达miR390植株侧根数增加 [16] 表 2 水稻中已克隆的根系生长发育相关激素基因及其功能
Table 2 Cloned genes and functions related to hormones of root growth and development
基因符号
Gene symbol基因号
Genomic Locus突变体表型
Mutant phenotype参考文献
ReferencesTDD1 Os04g0463500 ostdd1根长减短 [42] OsYUCCA1 Os01g0645400 osyucca1冠状根增加 [42] FIB Os01g0169800 突变体fib根系发育缺陷 [43] OsAUX1 Os01g0856500 osaux1根长增加,根毛较短,过表达植株相反 [44] OsPIN2 Os06g0660200 ospin2突变体根卷曲,侧根形成模式改变 [45] OsTIR1 Os05g0150500 ostir1初生根增长,不定根和侧根密度降低 [46] OsAFB2 Os04g0395600 osafb2初生根增长,不定根和侧根密度降低 [46] OsLOGL5 Os03g0857900 OsLOGL5表达量的改变影响根系伸长和侧根发育 [47] BG3 Os01g0680200 bg3-D根生长量减少 [48] OsBRD1 Os03g0602300 osbrd1根系发育不良,冠根数量少 [49] OsBRI1 Os01g0718300 osbri1根系生长异常 [50] OsBZR1 Os07g0580500 OsBZR1过表达植株侧根数增加 [49] OsKS1 Os04g0611800 ks1突变体根长较短和根分生组织更小 [51] OsSWEET3a Os05g0214300 OsSWEET3a的敲除系和过表达系均表现出根生长迟缓 [52] OsSLR1 Os03g0707600 突变体slr1根长较短且数目减少 [53] OsNCED2 Os12g0435200 干旱条件下,过表达植株冠根和侧根数增加,敲除突变体相反 [54] OsACS1 Os03g0727600 OsACS1过表达植株根系更短 [55] OsERS2 Os05g0155200 osers2突变体植株具有短根表型 [56] OsCOI2 Os03g0265500 MeJA处理后的coi2根比野生型长 [57] AIM1 Os02g0274100 突变体aim1根长和不定根根长显著变短 [58] -
[1] CHANG C R,HU Y B,SUN S B,et al. Proton pump OsA8 is linked to phosphorus uptake and translocation in rice[J]. Journal of Experimental Botany,2009,60(2) :557−565. DOI: 10.1093/jxb/ern298
[2] SUN Y F,LUO W Z,JAIN A,et al. OsPHR3 affects the traits governing nitrogen homeostasis in rice[J]. BMC Plant Biology,2018,18(1) :241. DOI: 10.1186/s12870-018-1462-7
[3] DAI X Y,WANG Y Y,ZHANG W H. OsWRKY74,a WRKY transcription factor,modulates tolerance to phosphate starvation in rice[J]. Journal of Experimental Botany,2016,67(3) :947−960. DOI: 10.1093/jxb/erv515
[4] LI K N,ZHANG S N,TANG S,et al. The rice transcription factor Nhd1 regulates root growth and nitrogen uptake by activating nitrogen transporters[J]. Plant Physiology,2022,189(3) :1608−1624. DOI: 10.1093/plphys/kiac178
[5] EL-KEREAMY A,BI Y M,MAHMOOD K,et al. Overexpression of the CC-type glutaredoxin,OsGRX6 affects hormone and nitrogen status in rice plants[J]. Frontiers in Plant Science,2015,6:934.
[6] CAO Y,YAN Y,ZHANG F,et al. Fine characterization of OsPHO2 knockout mutants reveals its key role in Pi utilization in rice[J]. Journal of Plant Physiology,2014,171(3/4) :340−348.
[7] ZHOU S L,JIANG W,LONG F,et al. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem[J]. The Plant Cell,2017,29(5) :1088−1104. DOI: 10.1105/tpc.16.00908
[8] SHI M,XIE Y R,ZHENG Y Y,et al. Oryza sativa actin-interacting protein 1 is required for rice growth by promoting actin turnover[J]. Plant Journal,2013,73(5) :747−760. DOI: 10.1111/tpj.12065
[9] XU L,WANG F,LI R L,et al. OsCYCP4s coordinate phosphate starvation signaling with cell cycle progression in rice[J]. Journal of Integrative Plant Biology,2020,62(7) :1017−1033. DOI: 10.1111/jipb.12885
[10] KIM C M,PARK S H,JE B I,et al. OsCSLD1,a cellulose synthase-like D1 gene,is required for root hair morphogenesis in rice[J]. Plant Physiology,2007,143(3) :1220−1230. DOI: 10.1104/pp.106.091546
[11] INUKAI Y,SAKAMOTO T,MORINAKA Y,et al. ROOT GROWTH INHIBITING,a rice endo-1,4-β-d-glucanase,regulates cell wall loosening and is essential for root elongation[J]. Journal of Plant Growth Regulation,2012,31(3) :373−381. DOI: 10.1007/s00344-011-9247-3
[12] XIONG G Y,LI R,QIAN Q,et al. The rice dynamin-related protein DRP2B mediates membrane trafficking,and thereby plays a critical role in secondary cell wall cellulose biosynthesis[J]. Plant Journal,2010,64(1) :56−70.
[13] QU L H,WU C Y,ZHANG F,et al. Rice putative methyltransferase gene OsTSD2 is required for root development involving pectin modification[J]. Journal of Experimental Botany,2016,67(18) :5349−5362. DOI: 10.1093/jxb/erw297
[14] PARK J J,YI J,YOON J,et al. OsPUB15,an E3 ubiquitin ligase,functions to reduce cellular oxidative stress during seedling establishment[J]. Plant Journal,2011,65(2) :194−205. DOI: 10.1111/j.1365-313X.2010.04416.x
[15] JOO J,LEE Y H,SONG S I. Rice CatA,CatB,and CatC are involved in environmental stress response,root growth,and photorespiration,respectively[J]. Journal of Plant Biology,2014,57(6) :375−382. DOI: 10.1007/s12374-014-0383-8
[16] LU Y Z,FENG Z,LIU X Y,et al. MiR393 and miR390 synergistically regulate lateral root growth in rice under different conditions[J]. BMC Plant Biology,2018,18(1) :261. DOI: 10.1186/s12870-018-1488-x
[17] MORITA S,NEMOTO K. Morphology and anatomy of rice roots with special reference to coordination in organo- and histogenesis[M]//Structure and Function of Roots. Dordrecht:Springer Netherlands,1995:75–86.
[18] SUN H W,LI W Q,BURRITT D J,et al. Strigolactones interact with other phytohormones to modulate plant root growth and development[J]. The Crop Journal,2022,10(6) :1517−1527. DOI: 10.1016/j.cj.2022.07.014
[19] ROBBINS N E II,DINNENY J R. Growth is required for perception of water availability to pattern root branches in plants[J]. Proceedings of the National Academy of Sciences of the United States of America,2018,115(4) :E822−E831.
[20] SEBASTIAN J,YEE M C,GOUDINHO VIANA W,et al. Grasses suppress shoot-borne roots to conserve water during drought[J]. Proceedings of the National Academy of Sciences of the United States of America,2016,113(31) :8861−8866.
[21] BENGOUGH A G,LOADES K,MCKENZIE B M. Root hairs aid soil penetration by anchoring the root surface to pore walls[J]. Journal of Experimental Botany,2016,67(4) :1071−1078. DOI: 10.1093/jxb/erv560
[22] CLARK L H,HARRIS W H. Observations on the root anatomy of rice (Oryza sativa L. ) [J]. American Journal of Botany,1981,68(2) :154. DOI: 10.1002/j.1537-2197.1981.tb12374.x
[23] MOTTE H,VANNESTE S,BEECKMAN T. Molecular and environmental regulation of root development[J]. Annual Review of Plant Biology,2019,70:465−488. DOI: 10.1146/annurev-arplant-050718-100423
[24] CARRILLO-CARRASCO V P,HERNANDEZ-GARCIA J,MUTTE S K,et al. The birth of a giant:Evolutionary insights into the origin of auxin responses in plants[J]. The EMBO Journal,2023,42(6) :e113018. DOI: 10.15252/embj.2022113018
[25] YAMAZAKI K,FUJIWARA T. The effect of phosphate on the activity and sensitivity of nutritropism toward ammonium in rice roots[J]. Plants,2022,11(6) :733. DOI: 10.3390/plants11060733
[26] WANG H Q,ZHAO X Y,XUAN W,et al. Rice roots avoid asymmetric heavy metal and salinity stress via an RBOH-ROS-auxin signaling cascade[J]. Molecular Plant,2023,16(10) :1678−1694. DOI: 10.1016/j.molp.2023.09.007
[27] ZHU S X,ZHAO W,SUN S X,et al. Community metagenomics reveals the processes of cadmium resistance regulated by microbial functions in soils with Oryza sativa root exudate input[J]. Science of the Total Environment,2024,949:175015. DOI: 10.1016/j.scitotenv.2024.175015
[28] WU B B,WANG J Y,DAI H Y,et al. Radial oxygen loss triggers diel fluctuation of cadmium dissolution in the rhizosphere of rice[J]. Environmental Science & Technology,2024,58(33) :14718−14725.
[29] HUANG Y Z,JI Z,TAO Y J,et al. Improving rice nitrogen-use efficiency by modulating a novel monouniquitination machinery for optimal root plasticity response to nitrogen[J]. Nature Plants,2023,9(11) :1902−1914. DOI: 10.1038/s41477-023-01533-7
[30] LEI Z L,DING Y X,XU W F,et al. Microbial community structure in rice rhizosheaths under drought stress[J]. Journal of Plant Ecology,2023,16(5) :rtad012. DOI: 10.1093/jpe/rtad012
[31] DING Z J,XU C,YAN J Y,et al. The LRR receptor-like kinase ALR1 is a plant aluminum ion sensor[J]. Cell Research,2024,34(4) :281−294. DOI: 10.1038/s41422-023-00915-y
[32] RONZAN M,PIACENTINI D,FATTORINI L,et al. Cadmium and arsenic affect root development in Oryza sativa L. negatively interacting with auxin[J]. Environmental and Experimental Botany,2018,151:64−75. DOI: 10.1016/j.envexpbot.2018.04.008
[33] QIN H,HUANG R F. The phytohormonal regulation of Na+/K+ and reactive oxygen species homeostasis in rice salt response[J]. Molecular Breeding,2020,40(5) :47. DOI: 10.1007/s11032-020-1100-6
[34] ARSOVA B,FOSTER K J,SHELDEN M C,et al. Dynamics in plant roots and shoots minimize stress,save energy and maintain water and nutrient uptake[J]. New Phytologist,2020,225(3) :1111−1119. DOI: 10.1111/nph.15955
[35] JIA Z T,GIEHL R F H,VON WIRÉN N. Local auxin biosynthesis acts downstream of brassinosteroids to trigger root foraging for nitrogen[J]. Nature Communications,2021,12(1) :5437. DOI: 10.1038/s41467-021-25250-x
[36] LI J H,ZHANG Z Y,CHONG K,et al. Chilling tolerance in rice:Past and present[J]. Journal of Plant Physiology,2022,268:153576. DOI: 10.1016/j.jplph.2021.153576
[37] GROVER M,BODHANKAR S,SHARMA A,et al. PGPR mediated alterations in root traits:Way toward sustainable crop production[J]. Frontiers in Sustainable Food Systems,2021,4:618230. DOI: 10.3389/fsufs.2020.618230
[38] WU Q Q,PENG X J,YANG M F,et al. Rhizobia promote the growth of rice shoots by targeting cell signaling,division and expansion[J]. Plant Molecular Biology,2018,97(6) :507−523. DOI: 10.1007/s11103-018-0756-3
[39] XU F Y,LIAO H P,ZHANG Y J,et al. Coordination of root auxin with the fungus Piriformospora indica and bacterium Bacillus cereus enhances rice rhizosheath formation under soil drying[J]. The ISME Journal,2022,16(3) :801−811. DOI: 10.1038/s41396-021-01133-3
[40] MEENA K K,BITLA U M,SORTY A M,et al. Mitigation of salinity stress in wheat seedlings due to the application of phytohormone-rich culture filtrate extract of methylotrophic actinobacterium Nocardioides sp. NIMMe6[J]. Frontiers in Microbiology,2020,11:2091.
[41] ZHANG Y J,DU H,XU F Y,et al. Root-bacteria associations boost rhizosheath formation in moderately dry soil through ethylene responses[J]. Plant Physiology,2020,183(2) :780−792. DOI: 10.1104/pp.19.01020
[42] ZHANG T,LI R N,XING J L,et al. The YUCCA-auxin-WOX11 module controls crown root development in rice[J]. Frontiers in Plant Science,2018,9:523. DOI: 10.3389/fpls.2018.00523
[43] YOSHIKAWA T,ITO M,SUMIKURA T,et al. The rice FISH BONE gene encodes a tryptophan aminotransferase,which affects pleiotropic auxin-related processes[J]. Plant Journal,2014,78(6) :927−936. DOI: 10.1111/tpj.12517
[44] SUN C D,LI D M,GAO Z Y,et al. OsRLR4 binds to the OsAUX1 promoter to negatively regulate primary root development in rice[J]. Journal of Integrative Plant Biology,2022,64(1) :118−134. DOI: 10.1111/jipb.13183
[45] LI W Q,ZHANG M J,QIAO L,et al. Characterization of wavy root 1,an agravitropism allele,reveals the functions of OsPIN2 in fine regulation of auxin transport and distribution and in ABA biosynthesis and response in rice (Oryza sativa L. ) [J]. The Crop Journal,2022,10(4) :980−992. DOI: 10.1016/j.cj.2021.12.004
[46] GUO F,HUANG Y Z,QI P P,et al. Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield,tillering,plant height,root system,germination,and auxinic herbicide resistance[J]. New Phytologist,2021,229(5) :2676−2692. DOI: 10.1111/nph.17061
[47] CHEN L,JAMESON G B,GUO Y C,et al. The LONELY GUY gene family:From mosses to wheat,the key to the formation of active cytokinins in plants[J]. Plant Biotechnology Journal,2022,20(4) :625−645. DOI: 10.1111/pbi.13783
[48] XIAO Y H,LIU D P,ZHANG G X,et al. Big Grain3,encoding a purine permease,regulates grain size via modulating cytokinin transport in rice[J]. Journal of Integrative Plant Biology,2019,61(5) :581−597. DOI: 10.1111/jipb.12727
[49] JIAO X M,WANG H C,YAN J J,et al. Promotion of BR biosynthesis by miR444 is required for ammonium-triggered inhibition of root growth[J]. Plant Physiology,2020,182(3) :1454−1466. DOI: 10.1104/pp.19.00190
[50] 赵雪松,王倩,闫青地,等. 油菜素内酯对水稻根系发育的调控作用[J]. 中国细胞生物学学报,2016,38(10) :1191−1198. DOI: 10.11844/cjcb.2016.10.0109 ZHAO X S,WANG Q,YAN Q D,et al. Function of brassinolide in the regulation of root development in rice[J]. Chinese Journal of Cell Biology,2016,38(10) :1191−1198. (in Chinese) DOI: 10.11844/cjcb.2016.10.0109
[51] HE Y Q,HONG G J,ZHANG H H,et al. The OsGSK2 kinase integrates brassinosteroid and jasmonic acid signaling by interacting with OsJAZ4[J]. The Plant Cell,2020,32(9) :2806−2822. DOI: 10.1105/tpc.19.00499
[52] MORII M,SUGIHARA A,TAKEHARA S,et al. The dual function of OsSWEET3a as a gibberellin and glucose transporter is important for young shoot development in rice[J]. Plant & Cell Physiology,2020,61(11) :1935−1945.
[53] DAVIÈRE J M,ACHARD P. A pivotal role of DELLAs in regulating multiple hormone signals[J]. Molecular Plant,2016,9(1) :10−20. DOI: 10.1016/j.molp.2015.09.011
[54] HUANG L Y,BAO Y C,QIN S W,et al. The ABA synthesis enzyme allele OsNCED2T promotes dryland adaptation in upland rice[J]. The Crop Journal,2024,12(1) :68−78. DOI: 10.1016/j.cj.2023.12.001
[55] QIN H,WANG J,CHEN X B,et al. Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress[J]. New Phytologist,2019,223(2) :798−813. DOI: 10.1111/nph.15824
[56] ZHAO H,DUAN K X,MA B,et al. Histidine kinase MHZ1/OsHK1 interacts with ethylene receptors to regulate root growth in rice[J]. Nature Communications,2020,11(1) :518. DOI: 10.1038/s41467-020-14313-0
[57] INAGAKI H,HAYASHI K,TAKAOKA Y,et al. Genome editing reveals both the crucial role of OsCOI2 in jasmonate signaling and the functional diversity of COI1 homologs in rice[J]. Plant & Cell Physiology,2023,64(4) :405−421.
[58] XU L,ZHAO H Y,RUAN W Y,et al. ABNORMAL INFLORESCENCE MERISTEM1 functions in salicylic acid biosynthesis to maintain proper reactive oxygen species levels for root meristem activity in rice[J]. The Plant Cell,2017,29(3) :560−574. DOI: 10.1105/tpc.16.00665
[59] WAADT R,SELLER C A,HSU P K,et al. Plant hormone regulation of abiotic stress responses[J]. Nature Reviews Molecular Cell Biology,2022,23(10) :680−694. DOI: 10.1038/s41580-022-00479-6
[60] ZHAO B Q,LIU Q Y,WANG B S,et al. Roles of phytohormones and their signaling pathways in leaf development and stress responses[J]. Journal of Agricultural and Food Chemistry,2021,69(12) :3566−3584. DOI: 10.1021/acs.jafc.0c07908
[61] MAO C J,HE J M,LIU L N,et al. OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development[J]. Plant Biotechnology Journal,2020,18(2) :429−442. DOI: 10.1111/pbi.13209
[62] ZHAO J,YANG B,LI W J,et al. A genome-wide association study reveals that the glucosyltransferase OsIAGLU regulates root growth in rice[J]. Journal of Experimental Botany,2021,72(4) :1119−1134. DOI: 10.1093/jxb/eraa512
[63] ZHANG S Z,WU T,LIU S J,et al. Disruption of OsARF19 is critical for floral organ development and plant architecture in rice (Oryza sativa L. ) [J]. Plant Molecular Biology Reporter,2016,34(4) :748−760. DOI: 10.1007/s11105-015-0962-y
[64] WANG M,QIAO J Y,YU C L,et al. The auxin influx carrier,OsAUX3,regulates rice root development and responses to aluminium stress[J]. Plant,Cell & Environment,2019,42(4) :1125–1138.
[65] YE R G,WU Y R,GAO Z Y,et al. Primary root and root hair development regulation by OsAUX4 and its participation in the phosphate starvation response[J]. Journal of Integrative Plant Biology,2021,63(8) :1555−1567. DOI: 10.1111/jipb.13142
[66] JIANG L H,YAO B L,ZHANG X Y,et al. Salicylic acid inhibits rice endocytic protein trafficking mediated by OsPIN3t and clathrin to affect root growth[J]. Plant Journal,2023,115(1) :155−174. DOI: 10.1111/tpj.16218
[67] GAO J,ZHAO Y,ZHAO Z K,et al. RRS1 shapes robust root system to enhance drought resistance in rice[J]. New Phytologist,2023,238(3) :1146−1162. DOI: 10.1111/nph.18775
[68] CHEN Y,YANG Q F,SANG S H,et al. Rice inositol polyphosphate kinase (OsIPK2) directly interacts with OsIAA11 to regulate lateral root formation[J]. Plant & Cell Physiology,2017,58(11) :1891−1900.
[69] KITOMI Y,INAHASHI H,TAKEHISA H,et al. OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice[J]. Plant Science,2012,190:116−122. DOI: 10.1016/j.plantsci.2012.04.005
[70] NI J,WANG G H,ZHU Z X,et al. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice[J]. Plant Journal,2011,68(3) :433−442. DOI: 10.1111/j.1365-313X.2011.04698.x
[71] QI Y H,WANG S K,SHEN C J,et al. OsARF12,a transcription activator on auxin response gene,regulates root elongation and affects iron accumulation in rice (Oryza sativa) [J]. New Phytologist,2012,193(1) :109−120. DOI: 10.1111/j.1469-8137.2011.03910.x
[72] WANG X F,HE F F,MA X X,et al. OsCAND1 is required for crown root emergence in rice[J]. Molecular Plant,2011,4(2) :289−299. DOI: 10.1093/mp/ssq068
[73] HAN Y F,ZHANG C Z,SHA H J,et al. Ubiquitin-conjugating enzyme OsUBC11 affects the development of roots via auxin pathway[J]. Rice,2023,16(1) :9. DOI: 10.1186/s12284-023-00626-3
[74] WYBOUW B,DE RYBEL B. Cytokinin - A developing story[J]. Trends in Plant Science,2019,24(2) :177−185. DOI: 10.1016/j.tplants.2018.10.012
[75] ZHAO J Z,YU N N,JU M,et al. ABC transporter OsABCG18 controls the shootward transport of cytokinins and grain yield in rice[J]. Journal of Experimental Botany,2019,70(21) :6277−6291. DOI: 10.1093/jxb/erz382
[76] GAO S P,FANG J,XU F,et al. CYTOKININ OXIDASE/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation[J]. Plant Physiology,2014,165(3) :1035−1046. DOI: 10.1104/pp.114.238584
[77] DO NASCIMENTO F C,DE SOUZA A F F,DE SOUZA V M,et al. OsCKX5 modulates root system morphology and increases nutrient uptake in rice[J]. Journal of Plant Growth Regulation,2022,41(6) :2157−2170. DOI: 10.1007/s00344-021-10419-x
[78] NONGPIUR R C,RAWAT N,SINGLA-PAREEK S L,et al. OsRR26,a type-B response regulator,modulates salinity tolerance in rice via phytohormone-mediated ROS accumulation in roots and influencing reproductive development[J]. Planta,2024,259(5) :96. DOI: 10.1007/s00425-024-04366-6
[79] LIU H L,HUANG J Q,ZHANG X J,et al. The RAC/ROP GTPase activator OsRopGEF10 functions in crown root development by regulating cytokinin signaling in rice[J]. The Plant Cell,2023,35(1) :453−468. DOI: 10.1093/plcell/koac297
[80] HOU J Q,ZHENG X K,REN R F,et al. The histone deacetylase 1/GSK3/SHAGGY-like kinase 2/BRASSINAZOLE-RESISTANT 1 module controls lateral root formation in rice[J]. Plant Physiology,2022,189(2) :858−873. DOI: 10.1093/plphys/kiac015
[81] JIANG Y H,BAO L,JEONG S Y,et al. XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice[J]. Plant Journal,2012,70(3) :398−408. DOI: 10.1111/j.1365-313X.2011.04877.x
[82] LO S F,YANG S Y,CHEN K T,et al. A novel class of gibberellin 2-oxidases control semidwarfism,tillering,and root development in rice[J]. The Plant Cell,2008,20(10) :2603−2618. DOI: 10.1105/tpc.108.060913
[83] LI J T,ZHAO Y,CHU H W,et al. SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation[J]. PLoS Genetics,2015,11(8) :e1005464. DOI: 10.1371/journal.pgen.1005464
[84] MO W P,TANG W J,DU Y X,et al. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 interaction controls seedling growth under salt stress[J]. Plant Physiology,2020,184(1) :506−517. DOI: 10.1104/pp.20.00024
[85] LI J N,ZHANG Y X,LI Z Y,et al. OsPEX1,an extensin-like protein,negatively regulates root growth in a gibberellin-mediated manner in rice[J]. Plant Molecular Biology,2023,112(1/2) :47−59.
[86] TENG Z N,LYU J H,CHEN Y K,et al. Effects of stress-induced ABA on root architecture development:Positive and negative actions[J]. The Crop Journal,2023,11(4) :1072−1079. DOI: 10.1016/j.cj.2023.06.007
[87] CHEN H,MA B,ZHOU Y,et al. E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein[J]. Proceedings of the National Academy of Sciences of the United States of America,2018,115(17) :4513−4518.
[88] SANTOSH KUMAR V V,YADAV S K,VERMA R K,et al. The abscisic acid receptor OsPYL6 confers drought tolerance to indica rice through dehydration avoidance and tolerance mechanisms[J]. Journal of Experimental Botany,2021,72(4) :1411−1431. DOI: 10.1093/jxb/eraa509
[89] XU N,CHU Y L,CHEN H L,et al. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging[J]. PLoS Genetics,2018,14(10) :e1007662. DOI: 10.1371/journal.pgen.1007662
[90] KAWAI T,SHIBATA K,AKAHOSHI R,et al. WUSCHEL-related homeobox family genes in rice control lateral root primordium size[J]. Proceedings of the National Academy of Sciences of the United States of America,2022,119(1) :e2101846119.
[91] YOON J,CHO L H,YANG W Z,et al. Homeobox transcription factor OsZHD2 promotes root meristem activity in rice by inducing ethylene biosynthesis[J]. Journal of Experimental Botany,2020,71(18) :5348−5364. DOI: 10.1093/jxb/eraa209
[92] MA B,YIN C C,HE S J,et al. Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L. ) seedlings[J]. PLoS Genetics,2014,10(10) :e1004701. DOI: 10.1371/journal.pgen.1004701
[93] ZHAO H,MA B,DUAN K X,et al. The GDSL lipase MHZ11 modulates ethylene signaling in rice roots[J]. The Plant Cell,2020,32(5) :1626−1643. DOI: 10.1105/tpc.19.00840
[94] MA B,ZHOU Y,CHEN H,et al. Membrane protein MHZ3 stabilizes OsEIN2 in rice by interacting with its Nramp-like domain[J]. Proceedings of the National Academy of Sciences of the United States of America,2018,115(10) :2520−2525.
[95] ZHOU Y,GAO Y H,ZHANG B C,et al. CELLULOSE SYNTHASE-LIKE C proteins modulate cell wall establishment during ethylene-mediated root growth inhibition in rice[J]. The Plant Cell,2024,36(9) :3751−3769. DOI: 10.1093/plcell/koae195
[96] WU F H,GAO Y P,YANG W J,et al. Biological functions of strigolactones and their crosstalk with other phytohormones[J]. Frontiers in Plant Science,2022,13:821563. DOI: 10.3389/fpls.2022.821563
[97] WANG B B,ZHU X L,GUO X L,et al. Nitrate modulates lateral root formation by regulating the auxin response and transport in rice[J]. Genes,2021,12(6) :850. DOI: 10.3390/genes12060850
[98] SUN H W,GUO X L,QI X J,et al. SPL14/17 act downstream of strigolactone signalling to modulate rice root elongation in response to nitrate supply[J]. Plant Journal,2021,106(3) :649−660. DOI: 10.1111/tpj.15188
[99] RAYA-GONZÁLEZ J,ORTIZ-CASTRO R,RUÍZ-HERRERA L F,et al. PHYTOCHROME AND FLOWERING TIME1/MEDIATOR25 regulates lateral root formation via auxin signaling in Arabidopsis[J]. Plant Physiology,2014,165(2) :880−894. DOI: 10.1104/pp.114.239806
[100] SUZUKI G,LUCOB-AGUSTIN N,KASHIHARA K,et al. Rice MEDIATOR25,OsMED25,is an essential subunit for jasmonate-mediated root development and OsMYC2-mediated leaf senescence[J]. Plant Science,2021,306:110853. DOI: 10.1016/j.plantsci.2021.110853
[101] MUZAFFAR A,CHEN Y S,LEE H T,et al. A newly evolved rice-specific gene JAUP1 regulates jasmonate biosynthesis and signalling to promote root development and multi-stress tolerance[J]. Plant Biotechnology Journal,2024,22(5) :1417−1432. DOI: 10.1111/pbi.14276
[102] WATERS M T,GUTJAHR C,BENNETT T,et al. Strigolactone signaling and evolution[J]. Annual Review of Plant Biology,2017,68:291−322. DOI: 10.1146/annurev-arplant-042916-040925
[103] XIANG D,MENG F N,WANG A D,et al. Root-secreted peptide OsPEP1 regulates primary root elongation in rice[J]. Plant Journal,2021,107(2) :480−492. DOI: 10.1111/tpj.15303
[104] LI J Y,MENG L J,REN S H,et al. OsGSTU17,a tau class glutathione S-transferase gene,positively regulates drought stress tolerance in Oryza sativa[J]. Plants,2023,12(17) :3166. DOI: 10.3390/plants12173166
[105] YANG H S,FANG Y Y,LIANG Z M,et al. Polyamines:Pleiotropic molecules regulating plant development and enhancing crop yield and quality[J]. Plant Biotechnology Journal,2024,22(11) :3194−3201. DOI: 10.1111/pbi.14440
[106] MAI H F,QIN T,WEI H,et al. Overexpression of OsACL5 triggers environmentally-dependent leaf rolling and reduces grain size in rice[J]. Plant Biotechnology Journal,2024,22(4) :833−847. DOI: 10.1111/pbi.14227
[107] GAO Y Q,GUO R,WANG H Y,et al. Melatonin increases root cell wall phosphorus reutilization via an NO dependent pathway in rice (Oryza sativa) [J]. Journal of Pineal Research,2024,76(5) :e12995. DOI: 10.1111/jpi.12995




 下载:
下载: