Research Progress on Effects of Plant Hormones and Other Regulatory Factors on Rice Root Growth and Development
-
摘要:
水稻(Oryza sativa L.)作为全世界重要的粮食作物,提高其产量是保障国家粮食安全的重要策略。根系作为水稻最重要的器官之一,在水稻的生长发育、信号感知、激素合成、水分和养分吸收以及产量形成中起重要作用。然而,水稻根系的形成及生长发育也受到多种因素的影响。其中,植物激素在调控水稻根系生长发育过程中发挥着重要作用,使植物能够根据环境需求调节其发育、生长和生理状态。本文综述了影响水稻根系生长发育的非生物因素和生物因素;概述了与根系生长相关的基因及调控网络,重点阐述了植物激素在水稻根系形成及生长发育中的作用机制;总结了几种激素之间相互协同对根系的生长调控,并对其他影响根系的小分子物质进行讨论,旨在为水稻根系研究及提高水稻产量提供理论依据,也为今后更加精细化研究激素信号通路之间复杂的串扰以及和其他信号之间的作用机制提供新方向。
Abstract:Being one of the major food sources in the world, rice (Oryza sativa L.) has been studied to promote its growth and increase its production for the security of human livelihood. Since the root system is crucial for the growth, signal perception, hormone synthesis, water and nutrient uptakes, and production of a rice plant, the formation, development, and functions of the system need to be well understood. However, since the affecting factors are numerous, the study could be challenging. For instance, plant hormones are of special interest regarding their roles in regulating root growth and development under ever-changing environmental conditions. This article comprehensively reviewed subjects involving abiotic and biotic factors, related genes and regulatory networks, regulating plant hormones, and synergisms among hormones, metabolic trace minerals, and low-molecular substances. By emphasizing the studies on the plant hormones, it is hoped that research to further rice yield improvement and in-depth understanding of the basic biological mechanisms, complex interactions/interferences among biochemicals, and functional signaling pathways associated with rice root system development and functions will be realized.
-
Keywords:
- rice /
- root structure /
- plant hormones /
- regulatory network /
- growth and development
-
0. 引言
【研究意义】党参(Codonopsis pilosula)为桔梗科(Campanulaceae)党参属(Codonopsis)植物,其干燥根性味甘平,归脾、肺经,具有健脾益肺、养血生津的作用[1],现代药理研究表明党参具有促进造血性能、调节血糖、抗炎、延缓衰老、抗缺氧和加强机体免疫力等多种功效[2]。目前,党参的年消耗量在5万t左右[3],其用量持续增加导致党参渣的大量产出,由于提取方式和加工工艺制约,党参渣无法得到很好利用,而直接废弃则会造成较大的资源浪费[4]。【前人研究进展】已有研究将党参渣用于饲料添加剂,可促进麻鸡生产性能的改善,并增加麻鸡养殖的经济效益[5];除此之外,有研究报道,废弃药渣(包括党参渣)可通过生物转化技术用于生产沼气,或利用热化学转化技术将其转换成燃料物质[6],或可通过微生物技术生产包含多种活性成分的制剂或新药[7]。因此,开展党参渣的资源性物质二次开发利用,提高党参渣利用率,促进党参产业持续高速发展,成为当前党参产业健康发展的重要一环[8]。近年来,随着国家减抗和替抗政策的陆续出台,微生物发酵中药渣的工艺在畜牧业领域的应用受到广为关注[9]。孟晓燕[10]经研究发现,发酵中药渣能提高牛血清中过氧化物歧化酶、血清总蛋白、谷胱甘肽和免疫球蛋白等血清指标,提升抗氧化能力。李华伟[11]经研究发现,饲喂3%发酵中药渣(当归Angelicae Sinensis Radix、黄芪Astragalus membranaceus、白芍Cynanchum otophyllum和熟地黄Rehmannia glutinosa)可增加断奶仔猪的平均日增重,提高生产性能。植物乳杆菌(Lactiplantibacillus plantarum)是一种重要的益生菌,具有改善肠道菌群功能、抑制有害微生物的作用,近几年被广泛应用于中药发酵[12−15],其产生的胞外多糖具有抗氧化活性[16]。Fu等[17]利用植物乳杆菌发酵中药材红景天,发现其抗氧化和抗光老化活性均明显提高。Eweys等[18]研究发现,利用植物乳杆菌发酵肉桂,不但能提高肉桂的抗氧化活性,而且肉桂中的没食子酸、对羟基苯甲酸、儿茶素和绿原酸等活性成分也有一定程度的增加。【本研究切入点】党参渣利用微生物技术处理是目前较常见的方式,但目前较少经植物乳杆菌进行发酵二次利用的报道[19−20]。【拟解决的关键问题】本研究首次采用植物乳杆菌发酵党参渣,并将代谢组学的方法应用于植物乳杆菌发酵党参渣的研究,分析经植物乳杆菌N-25发酵后党参渣中的代谢物成分以及代谢通路方面的变化,以期揭示植物乳杆菌N-25发酵对党参渣代谢的影响,为党参渣资源化利用提供科学依据。
1. 材料与方法
1.1 试验材料
1.1.1 党参渣及植物乳杆菌
党参渣(醇提来源,后经去醇、干燥)回收自福建九为生物科技有限公司。植物乳杆菌N-25(N-25)分离自宁夏泡菜,保存于本实验室。在前期研究发现,N-25菌株具有良好的生物学活性,用该菌株发酵饲料可释放更多的不饱和脂肪酸和必需氨基酸等物质,促进营养物质的消化吸收和改善肠道健康[21]。
1.1.2 试剂及仪器
甲醇(LC-MS级,CNW Technologies,上海),乙腈( LC-MS级,CNW Technologies,上海),超纯水(屈臣氏),乙酸(LC-MS级,SIGMA-ALDRICH,德国),异丙醇(LC-MS级,CNW Technologies,上海),超高效液相色谱仪(Vanquish,Thermo Fisher Scientific,美国),高分辨质谱仪(Orbitrap Exploris 120,Thermo Fisher Scientific,美国),离心机(Heraeus Fresco17,Thermo Fisher Scientific,美国),天平(BSA124S-CW,Sartorius,德国),超声仪(PS-60AL,雷德邦电子有限公司,深圳),匀浆机(JXFSTPRP-24,净信科技有限公司,上海),冷冻干燥机(LGJ-10C,四环福瑞科仪科技发展有限公司,北京)。
1.2 试验方法
1.2.1 植物乳杆菌的增殖
在烧杯中加入脑心浸出液肉汤(BHI)培养基38.5 g,加入
1000 mL蒸馏水后加热,冷却后于121 ℃高压灭菌15 min,放凉备用。冰箱内取出4 ℃保存的植物乳杆菌,以1∶10的比例加入以上BHI液体培养基中,充分摇匀,在35 ℃、100 r·min−1条件下于恒温摇床中培养24 h后,4 ℃冰箱存放备用。取扩增好的菌液作10倍递增稀释,得到菌液浓度为1×108 CFU·mL−1。1.2.2 党参渣发酵
称取干燥后的党参渣100.00 g于锥形瓶内,加入一定比例蒸馏水,于121 ℃高压蒸汽灭菌锅内灭菌20 min,待冷却至室温后,于超净工作台内接种1×108 CFU·mL−1 N-25,摇匀后放入恒温培养箱。参照秦楠等的植物乳杆菌发酵工艺(温度31 ~39 ℃、发酵时间2 ~10 d、接菌量0.5% ~4%、料液比12.5% ~25%)[22],以黄酮含量作为标准观察发酵效果,据此确定本研究的发酵工艺:发酵温度35 ℃,发酵时间2 d,含水量30%进行有氧发酵。无菌水作为空白对照组(Y1),低剂量N-25组(Y2)设置1%(菌液含量0.3×108 CFU·mL−1)接菌量;中剂量N-25组(Y3)设置3%(菌液含量0.9×108 CFU·mL−1)接菌量;高剂量N-25组(Y4)设置5%(菌液含量1.5×108 CFU·mL−1)接菌量。
1.2.3 党参渣发酵代谢物的提取
试验样本送至上海百趣生物医学科技有限公司进行提取并分析。将样品冷冻干燥后研磨,低温称取25 mg样品于EP管中,加入匀浆珠,后加入
1000 μL提取液(甲醇∶乙腈∶水=2∶2∶1,V/V,含同位素标记内标混合物);涡旋混匀30 s;放入匀浆仪中匀浆(35 Hz,4 min),再转移至冰水浴超声5 min,此步骤重复3次;−40 ℃静置1 h;将样品4 ℃、12000 r·min−1,离心15 min,取上清液;上清液于4 ℃、12000 r·min−1,离心15 min;取上清液上机检测;所有样品另取等量上清液混合成QC样品上机检测。1.2.4 上机检测
针对发酵中药有较多的非极性代谢物,本试验使用Vanquish超高效液相色谱仪,通过Phenomenex Kinetex C18(2.1 mm×50 mm,2.6 μm)液相色谱柱对目标化合物进行色谱分离,条件为:液相色谱A相为水相,含0.01%乙酸,B相为异丙醇∶乙腈(1∶1,V/V)。样品盘温度:4 ℃,进样体积:2 μL。使用Xcalibur软件分析一级、二级质谱数据。
1.3 数据统计与分析
原始数据经ProteoWizard软件转成mzXML格式后,使用R包进行代谢物鉴定,所用数据库为BiotreeDB(V3.0),用R包进行可视化,采用Ropls进行主成分分析(principal component analysis, PCA)、正交偏最小二乘判别分析(orthogonal partialleast squares discriminant analysis, OPLS-DA);采用MetaboAnalyst对差异代谢物进行京都基因与基因组百科全书(Kyoto encyclopediaof genes and genomes, KEGG)代谢通路富集分析。基于SPSS26.0对相关组学数据进行单因素方差分析及差异显著性检验,使用Adobe Photoshop 2022进行绘图。
2. 结果与分析
2.1 质控分析
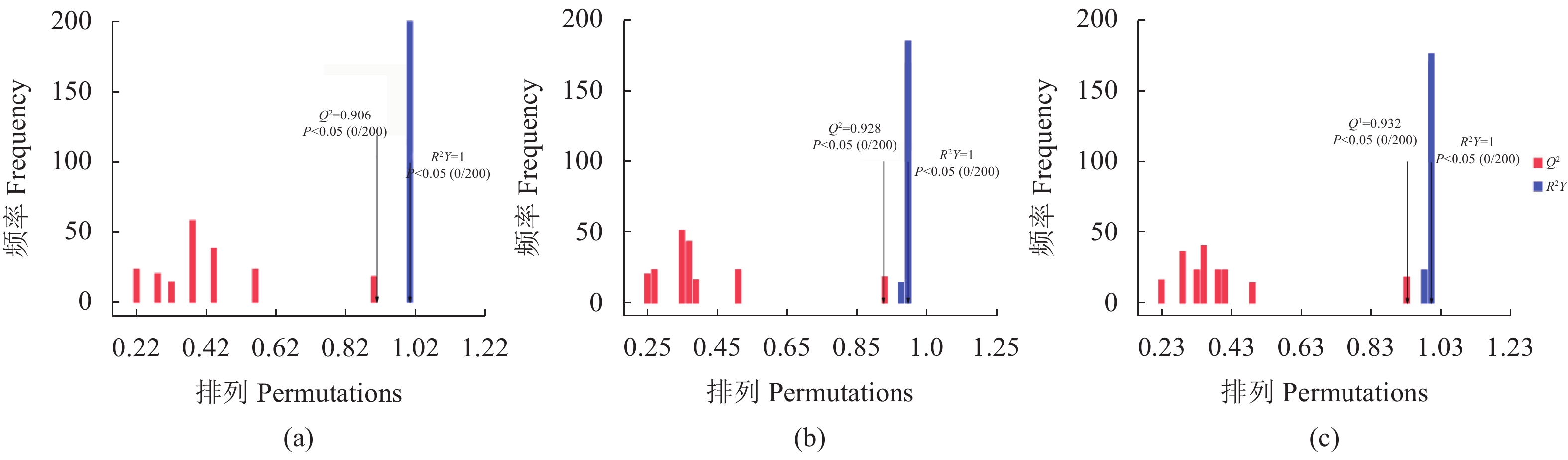
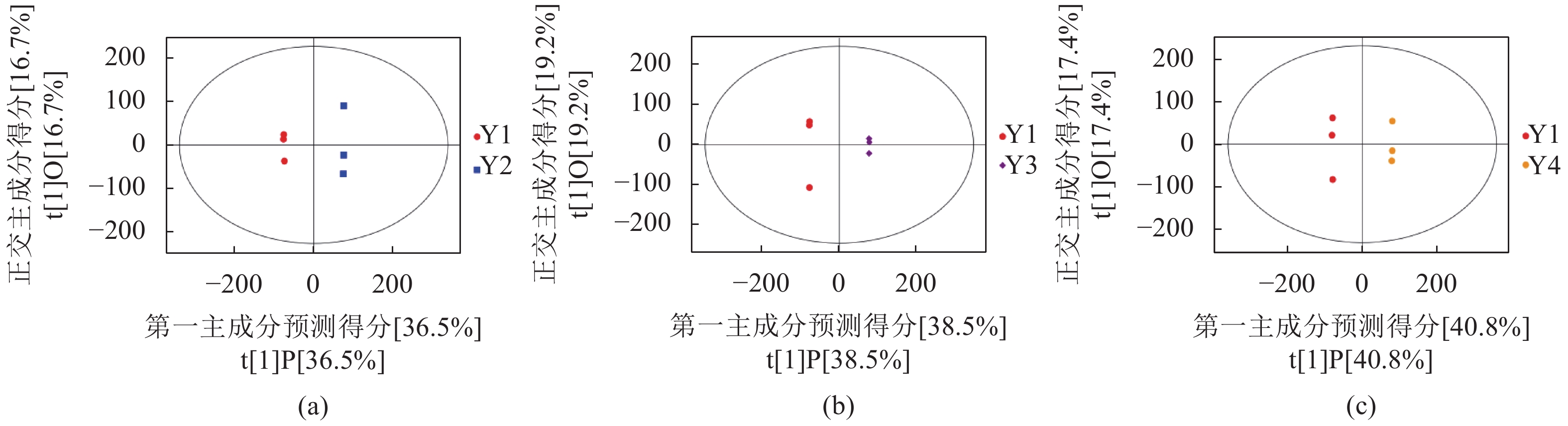
从图1中可知试验组(Y2、Y3和Y4)与Y1在OPLS-DA得分图上样本间离散程度不明显,均匀地分布于PC1左右两侧,说明组间差异小,组间整体代谢水平差异较大。通过随机改变分类变量Y的排列顺序,多次(次数n=200)建立对应的OPLS-DA模型以获取随机模型的R和Q值,来防止OPLS-DA模型过拟合并考察模型的质量,由图2可知Y1 vs. Y2的R2截距为0.99,Q2截距为−0.04;Y1 vs. Y2的R2截距为0.99,Q2截距为−0.09;Y1 vs. Y3的R2截距为0.99,Q2截距为−0.12,说明这3组OPLS-DA模型可靠。因此后续差异代谢物筛选可依据OPLS-DA模型。
2.2 代谢物分类统计
原始数据包含3个质控样本和12个试验样本。Y1组中鉴定到
2710 个代谢物,经N-25发酵后,试验组共鉴定到2736 种代谢物。发酵产物中的代谢物质如表1所示,其中主要的代谢物有脂类和类脂分子(包括柠檬酸、棕榈酸、月桂酸、γ-亚麻酸、黄芪皂苷Ⅲ、亚麻酸、丙戊酸、亚油酸、黄芪皂苷Ⅱ等代谢物),占比4.349%;莽草酸和苯丙烷类(包括异银杏双黄酮、去甲基川陈皮素、桑黄酮 E等黄酮类物质;阿魏内酯A、佛手苷内酯、异紫花前胡内酯等香豆素类代谢物;咖啡酸、肉桂酸苄酯、柠檬酸B、荜澄茄脂酮等木脂素及其他代谢物),占比4.094%;苯丙烷类和聚酮(包括川陈皮素、黄芩苷、香叶木素、7,8-二羟基黄酮等黄酮类物质;异黄酮类及其他代谢物),占比3.509%;有机杂环化合物(包括吡啶及衍生物、吲哚和衍生物、哌啶、吡喃、呋喃并吡喃类、喹啉及衍生物、吲哚和衍生物等物质),占比3.253%;苯类化合物(包括异香兰素、4-羟基扁桃酸、香草醛、地骨皮乙素等酚类物质;水杨酸、苯甲酸、邻乙酰氨基酚等苯环型化合物;萘类、蒽类和酚酯类物质),占比3.070%。表 1 代谢物分类及占比Table 1. Proportions of classified metabolites代谢物种类
Metabolite class数量
Number占比
Percentage/%脂质和类脂分子
Lipids and lipid-like molecules119 4.349 莽草酸和苯丙烷类
Shikimates and Phenylpropanoids112 4.094 苯丙烷和聚酮
Phenylpropanoids and polyketides96 3.509 有机杂环化合物
Organoheterocyclic compounds89 3.253 苯环型化合物
Benzenoids84 3.070 脂肪酸
Fatty acids68 2.485 有机酸及其衍生物
Organic acids and derivatives65 2.376 萜类化合物
Terpenoids65 2.376 生物碱类
Alkaloids59 2.156 有机含氧化合物
Organic oxygen compounds44 1.608 氨基酸和肽核苷
Amino acids and Peptides32 1.170 核苷酸和类似物
Nucleosides, nucleotides, and analogues29 1.060 碳水化合物
Carbohydrates19 0.694 聚酮
Polyketides16 0.585 生物碱及其衍生物
Alkaloids and derivatives15 0.548 有机氮化合物
Organic nitrogen compounds9 0.329 木脂素、新木脂素及相关化合物
Lignans, neolignans and related compounds5 0.183 其他 Others 1810 66.155 共计 Total 2736 100.000 2.3 N-25发酵前后差异代谢物筛选
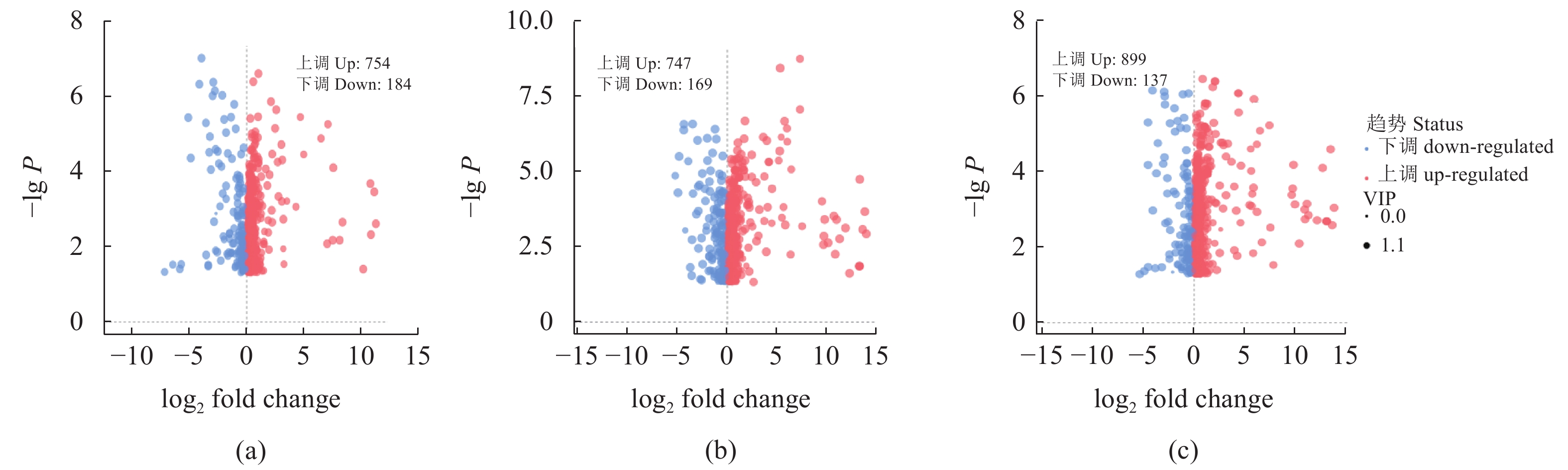
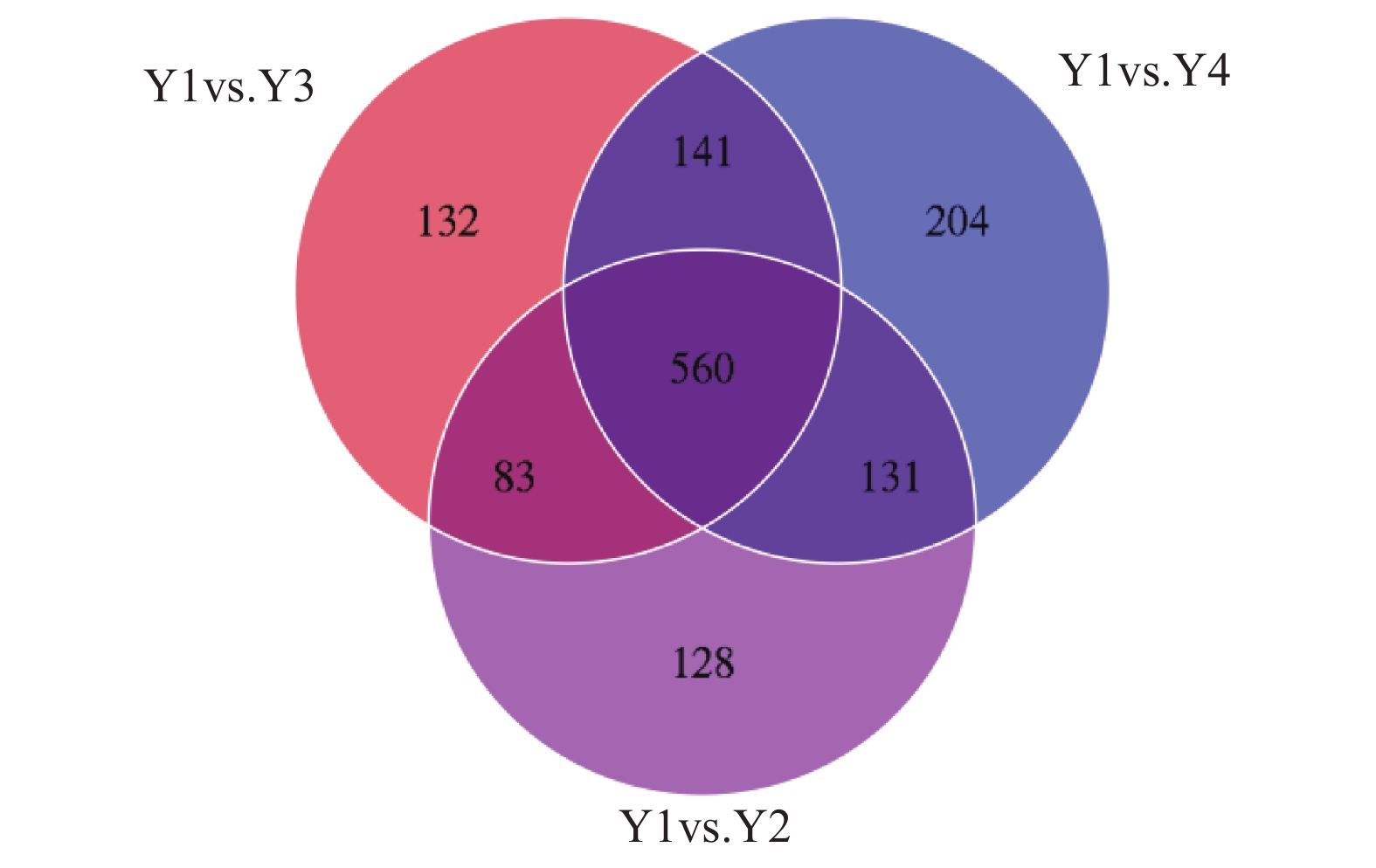
根据OPLS-DA结果,以VIP>1且P<0.05作为筛选条件,如图3所示,Y2比Y1组得到938个标志性差异代谢物,其中上调的有754个,下调的有184个;Y3比Y1组得到916个标志性差异代谢物,其中上调的有747个,下调的有169个;Y4比Y1组得到1036个标志性差异代谢物,其中上调的有899个,下调的有137个。图4展示了各组间差异代谢物之间的关系,三组间共同的差异代谢物有560个,Y2组特有差异代谢物为128种,包括3-氰基-L-丙氨酸、丙氨酸、2-萘酚、甘氨酸、甘露三糖、10-羟基癸酸等,Y3组有132种特有差异代谢物,包括康柠酸、月桂酸、3-羟基-3-甲基-2-氧代戊酸酯、异苹果酸、2-乙酰乳酸、L-亮氨酸等;Y4组特有的差异代谢物有204种,包括2-异丙基苹果酸、3-异丙基苹果酸、丙酮酸、L-异亮氨酸、L-缬氨酸等。
![]() 图 3 Y1组对Y2组、Y1组对Y3组、Y1组对Y4组的差异代谢物筛选火山图(a)Y1 vs.Y2;(b)Y1vs.Y3;(c)Y1vs.Y4。横坐标代表该组对比各物质的倍数变化(取以2为底的对数),纵坐标表示 t 检验的 P值(取以 10 为底对数的负数)。Figure 3. Volcanic diagram of differential metabolite screening between Y1 and Y2, Y1 and Y3, and Y1 and Y4(a) Y1 vs. Y2; (b) Y1 vs. Y3; (c) Y1 vs. Y4; horizontal axis: comparison of objects in same group on multiple quality changes (value in log2); vertical axis: P-values in student t-test (negative value of log10).
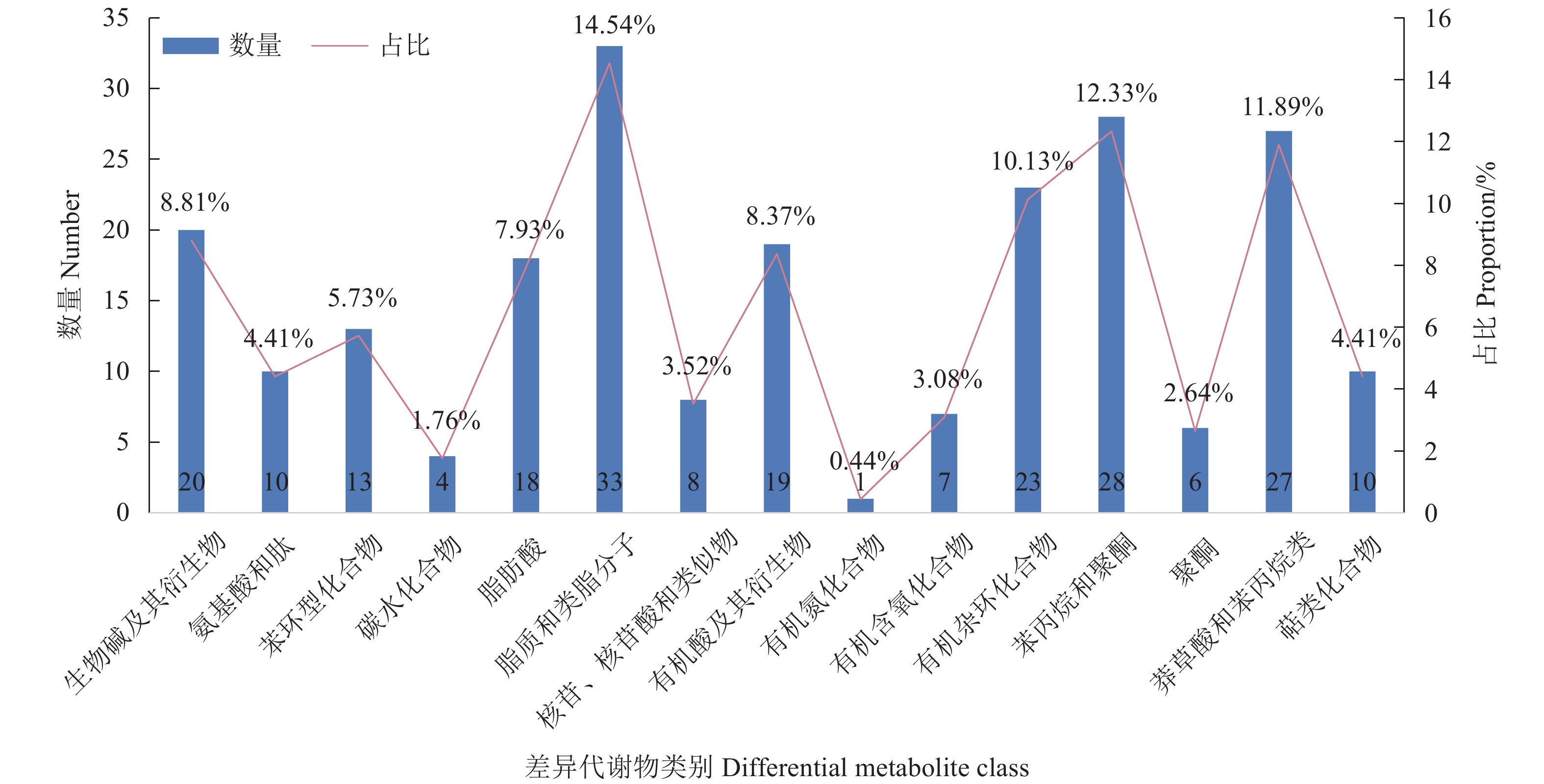
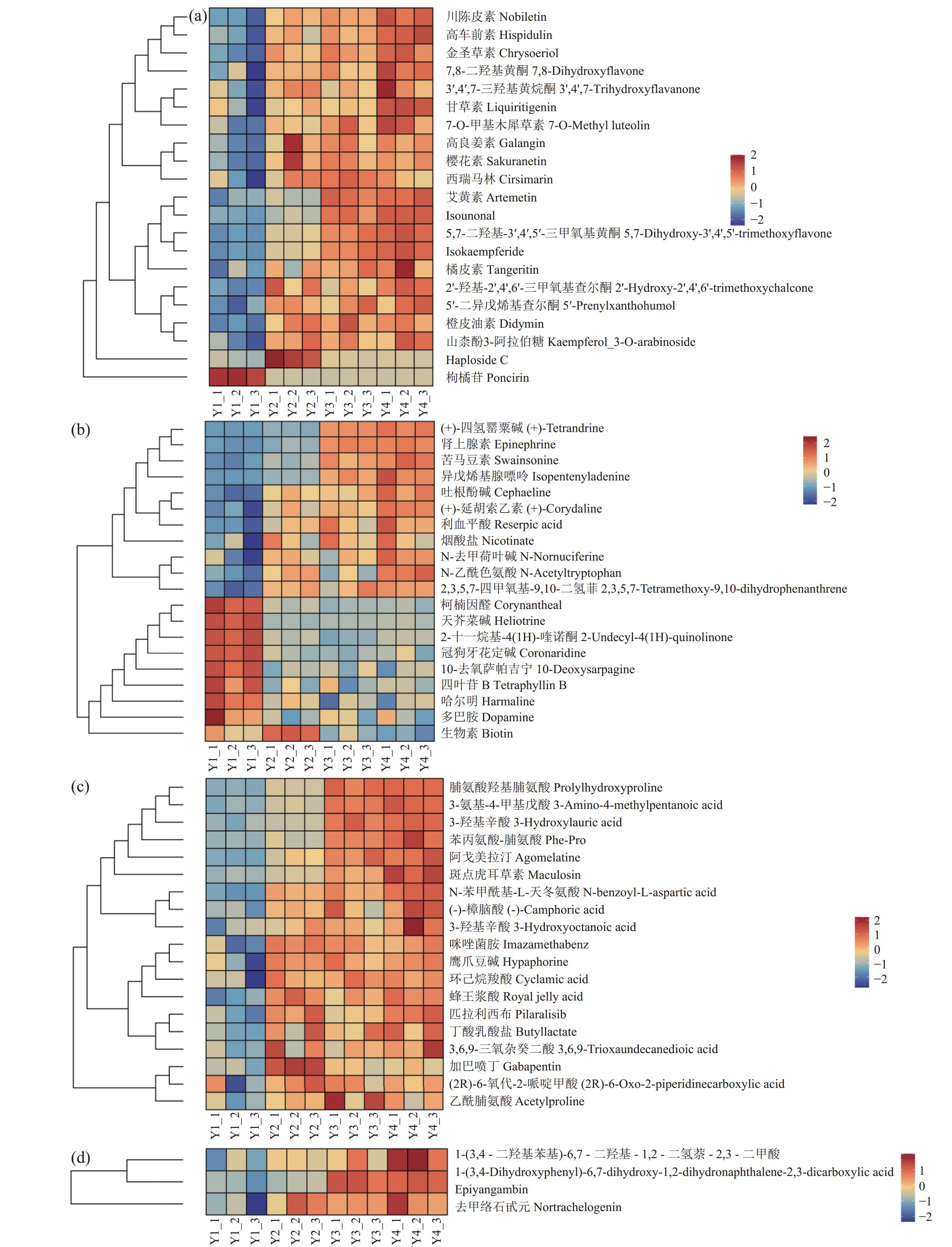
图 3 Y1组对Y2组、Y1组对Y3组、Y1组对Y4组的差异代谢物筛选火山图(a)Y1 vs.Y2;(b)Y1vs.Y3;(c)Y1vs.Y4。横坐标代表该组对比各物质的倍数变化(取以2为底的对数),纵坐标表示 t 检验的 P值(取以 10 为底对数的负数)。Figure 3. Volcanic diagram of differential metabolite screening between Y1 and Y2, Y1 and Y3, and Y1 and Y4(a) Y1 vs. Y2; (b) Y1 vs. Y3; (c) Y1 vs. Y4; horizontal axis: comparison of objects in same group on multiple quality changes (value in log2); vertical axis: P-values in student t-test (negative value of log10).为探究N-25对党参渣成分的影响,试验选取3个组共有的差异代谢物进行分析,其中被注释的差异代谢物有227个,图5展示了主要差异代谢物的分类与占比,可观察到主要差异代谢物有脂质和类脂分子、苯丙素烷和聚酮化合物、莽草酸和苯丙烷类、有机杂环化合物、生物碱及其衍生物、有机酸及衍生物以及脂肪酸。为了进一步观察植物乳杆菌N-25发酵引起的活性成分的变化,分别制作黄酮类化合物、木脂素、生物碱及其衍生物、有机酸及其衍生物的热图(图6)。
如图6所示,经N-25发酵后(图6),鉴定到川陈皮素、7-O-甲基木樨草素、高车前素、金圣草素和7,8-二羟基黄酮等18种黄酮类化合物含量均增加,较对照含量增加1.13倍;注释到的生物碱共20种,其中苦马豆素、烟酸盐、肾上腺素、(+)-四氢罂粟碱、异戊烯基腺嘌呤、吐根酚碱、(+)-延胡索乙素、利血平酸含量较对照增加1.01倍;鉴定到的3-羟基月桂酸、蜂王浆酸和(-)-樟脑酸等19种有机酸及其衍生物的相对含量较对照组增加1.16倍;另外,Epiyangambin、去甲络石甙元和1-(3,4-二羟基苯基)-6,7-二羟基-1,2-二氢萘-2,3-二甲酸3种木脂素在发酵后的党参渣中含量较对照增加1.04倍。
2.4 KEGG代谢通路分析
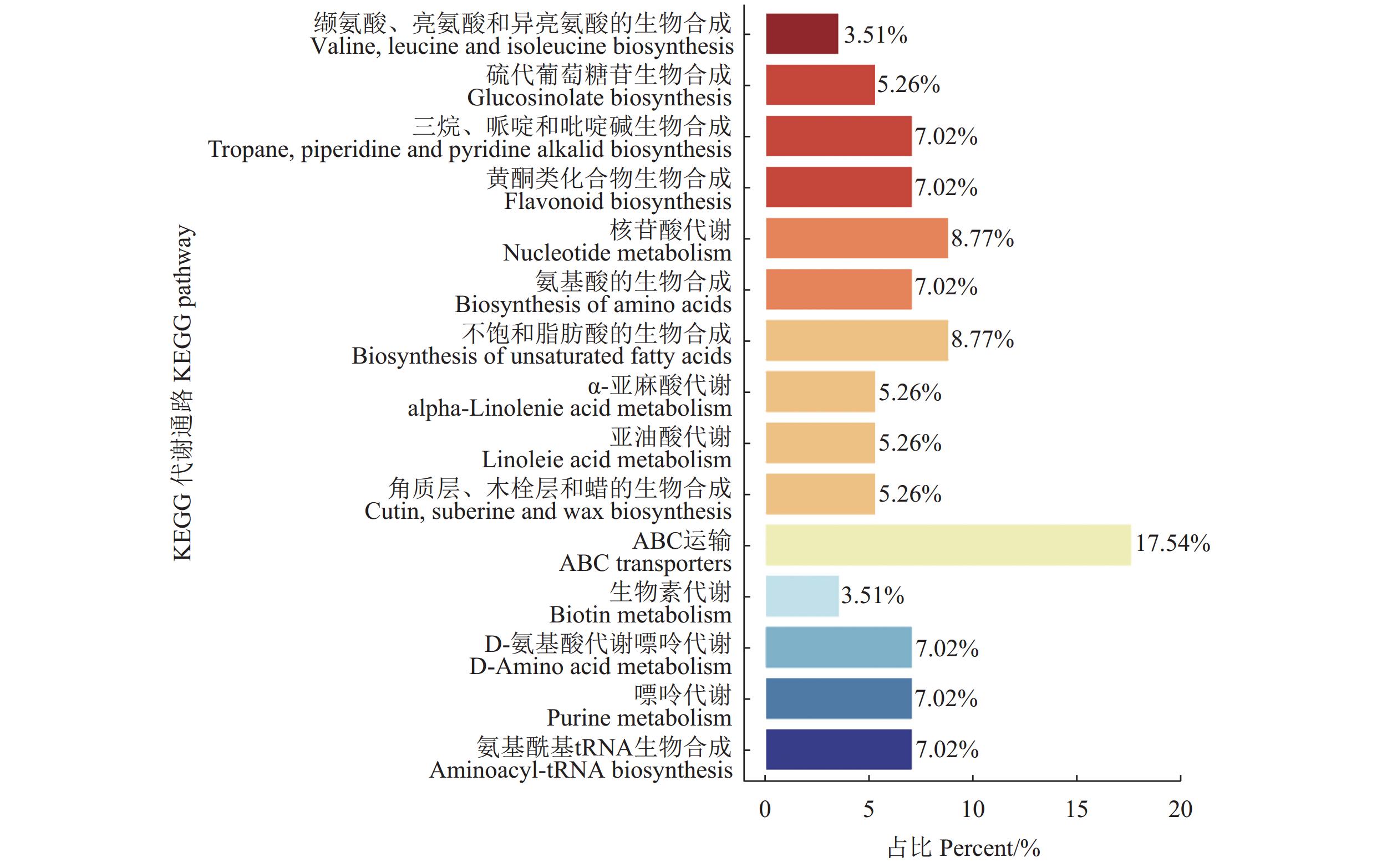
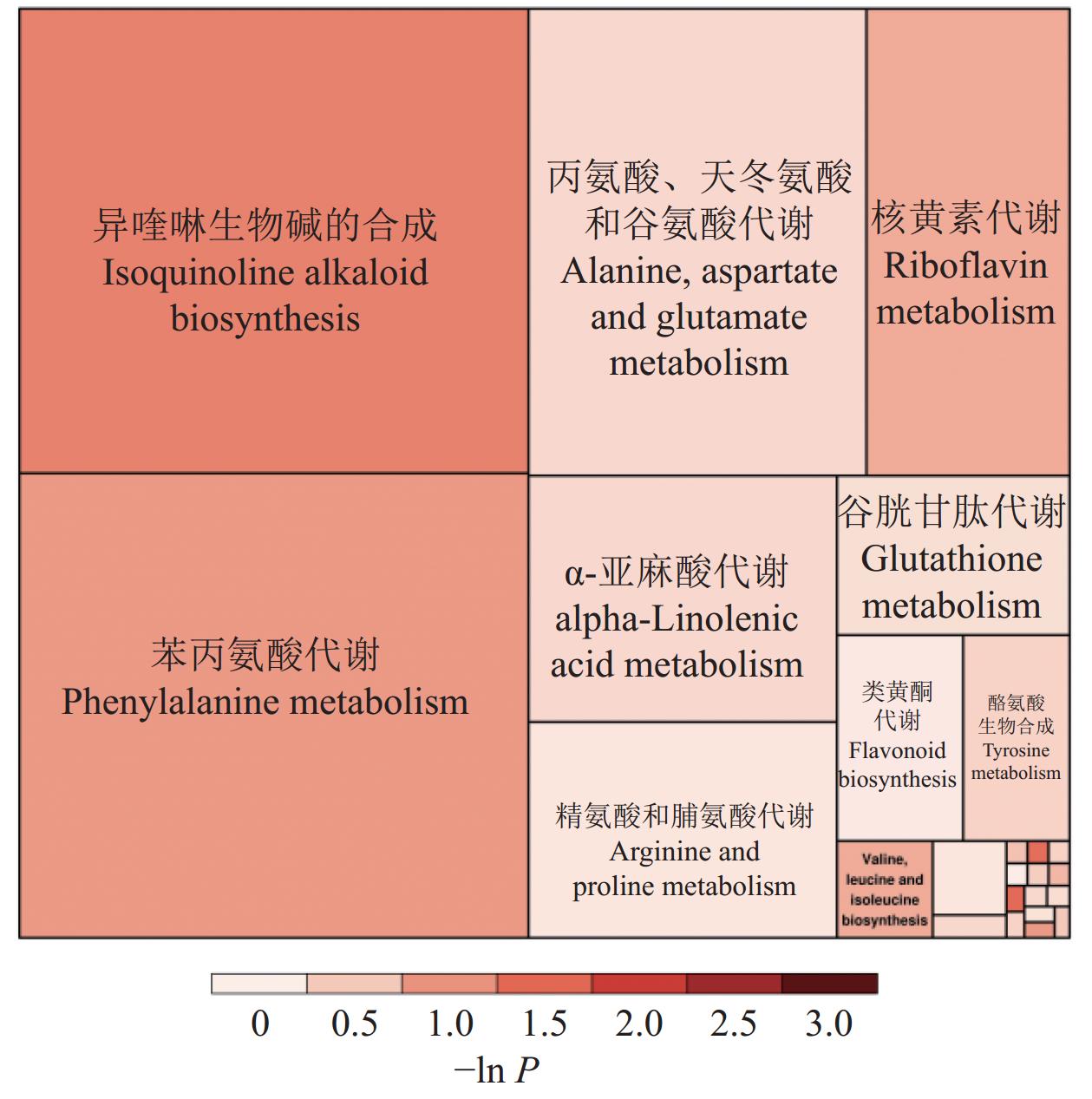
将筛选到的三组间共同的差异代谢物与KEGG数据库比对并进行通路富集分析。结果表明,在筛选出的560种具有显著差异代谢物中被KEGG注释到的共有56条通路。图7展示了差异代谢物富集到的部分通路,其中20.33%差异代谢物富集在代谢通路,13.19%差异代谢物富集在次级代谢物的生成通路,5.5%富集在ABC运输通路,3.3%富集在辅因子的生物合成通路,2.7%富集在核苷酸代谢,2.7%富集在不饱和脂肪酸的生成通路。其中关键通路包括异喹啉生物碱生物合成通路,苯丙氨酸代谢通路,核黄素代谢,丙氨酸、天冬氨酸和谷氨酸代谢通路,α-亚麻酸代谢通路(图8)。
2.5 特有差异代谢物KEGG通路分析
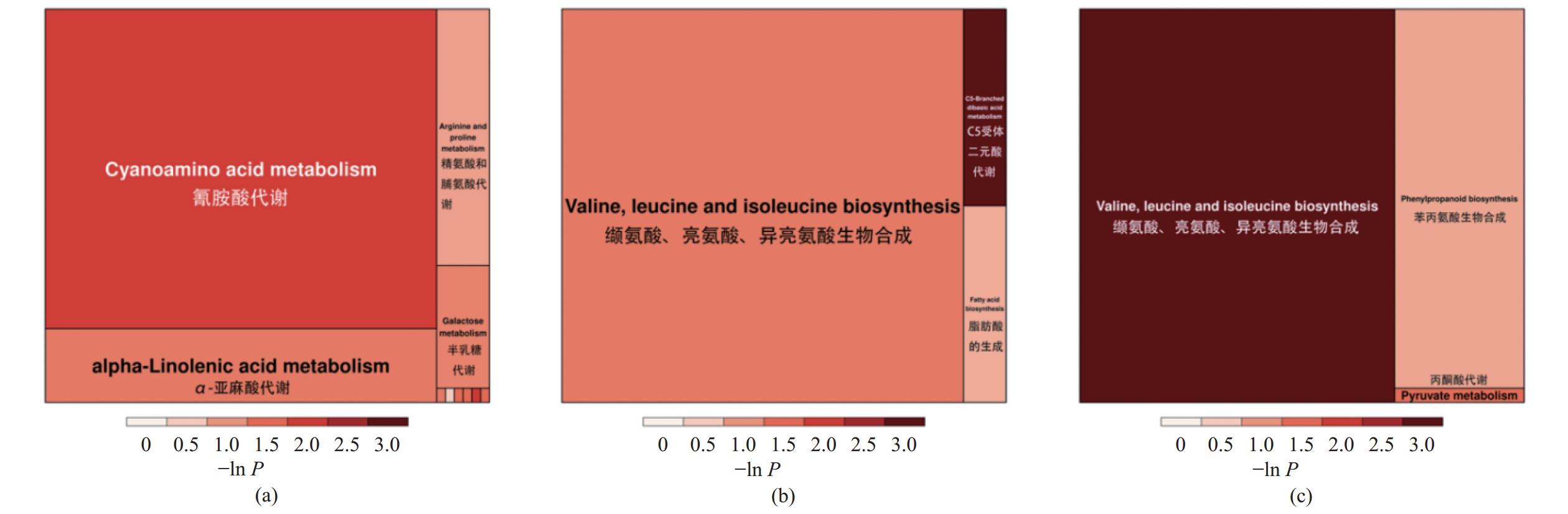
如图9所示,1%N-25发酵党参渣组,其代谢物富集在ABC运输、丁酸盐代谢、烟酸和烟酰胺代谢通路上;3%N-25发酵党参组,其代谢物富集在C5受体二元酸代谢、缬氨酸、亮氨酸、异亮氨酸生物合成、丙氨酸、天冬氨酸和谷氨酸代谢通路上;5%N-25发酵党参组,其代谢物富集在次生代谢物的生物合成、咖啡因代谢、缬氨酸、亮氨酸、异亮氨酸生物合成代谢通路上。
为进一步研究不同接菌量的N-25发酵对党参渣代谢物及其相关代谢通路的影响,通过对差异代谢物所在通路的综合分析(包括富集分析和拓扑分析),对通路进行进一步的筛选,三组差异代谢物相关性最高的关键通路如图10所示,Y1 vs.Y2特异性差异代谢物集中的关键通路主要为氰氨酸代谢通路,Y1 vs. Y3特异性差异代谢物集中的关键通路主要为缬氨酸、亮氨酸、异亮氨酸生物合成,Y1 vs. Y4特异性差异代谢物集中的关键通路主要为缬氨酸、亮氨酸、异亮氨酸生物合成。
3. 讨论与结论
本研究以N-25为发酵菌株,对党参渣进行单菌发酵,基于UHPLC-QE-MS的非靶代谢组学对发酵后的党参渣进行分析,结果显示,经过N-25发酵后,发酵产物共鉴定出
2737 种代谢物,包括32种羧酸及其衍生物,44种有机含氧化合物,55种脂肪酰基,70种黄酮类物质,74种生物碱,65种萜类,68种脂肪酸,32种氨基酸等。Zeng等[23]在废弃党参中共鉴定出1508 个代谢物,463种被鉴定为DEMs,可分为42种羧酸及其衍生物、34种类固醇和衍生物甾体、33种有机含氧化合物、32种脂肪酰基、21种黄酮类和其他类、26种生物碱、12种萜类和其他化合物。相较前人研究,发酵产物中活性成分的种类和含量都有增加,表明N-25发酵能提高党参渣的活性物质的释放,对党参渣的资源化利用提供了一种可能的有效途径。贾旭森[24]利用酵母菌发酵党参渣,发现黄酮、三萜、氨基酸含量等均有不同程度的增加。本研究采用N-25发酵党参渣,其黄酮、三萜、氨基酸含量显著增加,且与前人相比,增加幅度更明显,表明N-25对党参渣的发酵展现出了更优异的效果。本研究中含量增加的黄酮化合物,如川陈皮素和木樨草素具有良好的抗炎抗肿瘤作用[25−27],提示黄酮化合物的积累可能与N-25发酵党参渣中抗炎效应存在潜在关联,但还需进一步验证。苦马豆素作为一种生物碱,具有抗病毒和抗肿瘤的作用[28];烟酸盐具有抗炎作用[29];Epiyangambin具有显著的降压作用,并在预防和治疗血栓形成方面具有潜在应用价值[30−31];去甲络石甙元有抗氧化、抗炎和抗癌作用[32−33];3-羟基月桂酸和蜂王浆酸已被证实具有抗炎活性[34−36],(-)-樟脑酸可显著抑制前列腺素E2(PGE2)、白三烯B4(LTB4)及IL-6等炎症介质的生物合成[37]。本研究中,以上活性物质的含量均明显增加,进一步表明N25发酵能有效提升党参渣中活性成分的相对含量。
本研究中N-25发酵党参渣涉及众多通路,包括异喹啉生物碱合成通路、苯丙氨酸代谢通路、α-亚麻酸代谢通路等,其中相关性最强的是异喹啉生物碱合成通路,发酵后党参渣中异喹啉生物碱合成相关代谢物(+)-延胡索乙素、(+)-粉防己碱、吐根酚碱含量明显增加,多巴胺含量明显下降,推测N-25发酵的作用下存在N-甲基转移酶催化作用和伯胺N-甲基转移酶(NCS)催化作用,酪氨酸脱羧酶(TYDC)脱羧作用,将多巴胺转变为不同的异喹啉生物碱[38]。因异喹啉生物碱具有良好的抗炎作用[39−40],故认为N-25发酵党参渣对异喹啉生物碱活性物质的生成有积极的作用。
本试验研究结果表明,不同剂量N-25可以诱导党参渣产生不同特定功能的代谢产物,并且这些产物所涉及的功能也有所不同。Y2组中关键的特有代谢物3-氰基-L-丙氨酸与关键通路氰氨酸代谢通路直接相关[41−42],起到对发酵产物的解毒作用。亮氨酸、异亮氨酸和缬氨酸是人体营养必需支链氨基酸,它们不仅是蛋白质的基本组成单位,而且在细胞信号和分子调节、糖脂代谢、凋亡和自噬等方面发挥着重要的生理作用[43];Y3组主要特有代谢物2-Methyl-maleate参与关键通路为亮氨酸、异亮氨酸和缬氨酸合成通路,且Y3组亮氨酸和缬氨酸含量显著上升;Y4组关键的特有代谢物3-异丙基苹果酸和2-异丙基苹果酸是其关键通路亮氨酸、异亮氨酸和缬氨酸合成通路途径中的重要中间体[44−45];Y4组中亮氨酸和缬氨酸含量高于Y3组。因此,不同的N-25接菌量会产生不同的活性物质,但其作用机制还需进一步挖掘。
综上,植物乳杆菌N-25发酵党参渣,促进了黄酮类化合物、木脂素、生物碱及其衍生物、有机酸及其衍生物等活性物质的生成与释放;其共有差异代谢物富集的显著性差异代谢通路为异喹啉生物碱生物合成。
-
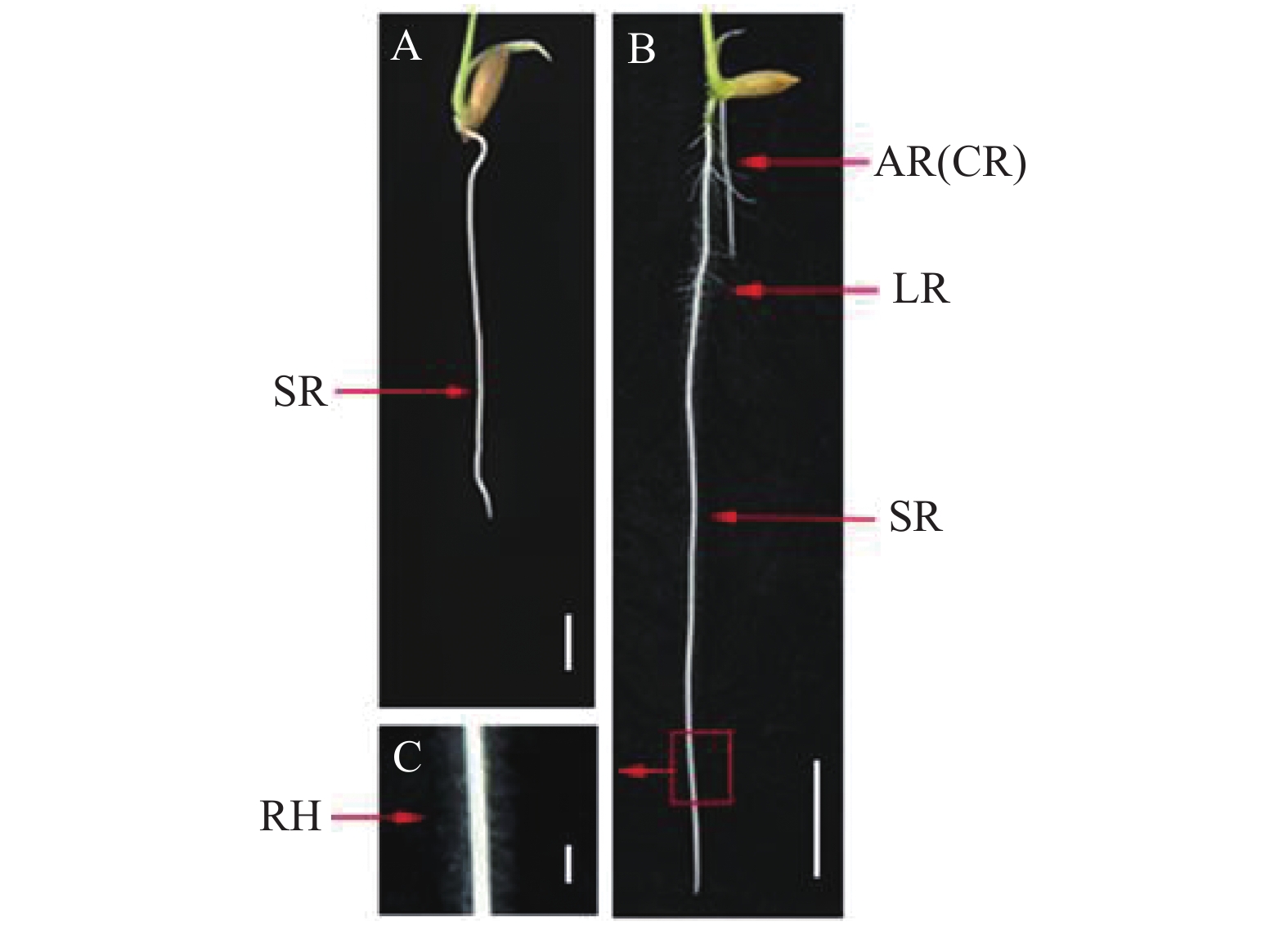
图 1 水稻根系组成[18]
A:4日龄水稻幼苗的根系, 标尺= 0.5 cm;B:7日龄水稻幼苗的根系,标尺= 1 cm;C:水稻根毛,标尺= 1 mm。SR:初生根;AR: 不定根;CR:冠根;RH:根毛。
Figure 1. Root system of a rice plant [18]
A: root of a 4-d-old rice seeding, scale bar=0.5 cm; B: root of a 7-d-old rice seeding, scale bar=1 cm; C: root hairs of a rice plant, scale bar=1 mm; SR: seminal root; AR: adventitious root; CR: crown root; RH: root hair.
表 1 水稻中已克隆的根系生长发育相关基因及其功能
Table 1 Cloned genes and functions associated with rice root growth and development
基因符号
Gene symbol基因号
Genomic Locus突变体表型
Mutant phenotype参考文献
ReferencesOsA8 Os03g0100800 OsA8的敲除突变植株根系生物量减少 [1] OsPHR3 Os02g0139000 OsPHR3突变体不定根和根毛发育减弱 [2] OsWRKY74 Os09g0334500 过表达植株在Pi或是P缺乏时,根的生物量更高 [3] OsAMT1;3 Os02g0620500 低铵条件下,敲除突变体植株总侧根数、种子根和侧根长度均降低 [4] OsGRX6 Os01g0667900 过表达OsGRX6的植株根干重下降 [5] OsPHO2 Os05g0557700 OsPHO2的敲除突变体植株根系更短 [6] GCN5 Os10g0415900 GCN5敲除突变体植株冠状根较少,根长降低 [7] OsAIP1 Os01g0125800 OsAIP1过表达和敲除植株根毛长度较短,直径增大 [8] OsCYCP4;1 Os10g0563900 过表达OsCYCP4;1植株根长度较短 [9] OsCSLD1 Os10g0578200 OsCSLD1突变体植株根毛长度偏短、局部扭曲膨胀 [10] RT Os04g0497200 突变体rt根长较短 [11] OsDRP2B Os02g0738900 突变体主根长度变短,侧根直径增加 [12] OsTSD2 Os02g0755000 突变体初生根较短、侧根更密 [13] OsPUB15 Os08g0110500 OsPUB15突变体种子根缺失 [14] OsCatB Os06g0727200 过表达CatB植株根重下降 [15] miR390 Os03g0817300 过表达miR390植株侧根数增加 [16] 表 2 水稻中已克隆的根系生长发育相关激素基因及其功能
Table 2 Cloned genes and functions related to hormones of root growth and development
基因符号
Gene symbol基因号
Genomic Locus突变体表型
Mutant phenotype参考文献
ReferencesTDD1 Os04g0463500 ostdd1根长减短 [42] OsYUCCA1 Os01g0645400 osyucca1冠状根增加 [42] FIB Os01g0169800 突变体fib根系发育缺陷 [43] OsAUX1 Os01g0856500 osaux1根长增加,根毛较短,过表达植株相反 [44] OsPIN2 Os06g0660200 ospin2突变体根卷曲,侧根形成模式改变 [45] OsTIR1 Os05g0150500 ostir1初生根增长,不定根和侧根密度降低 [46] OsAFB2 Os04g0395600 osafb2初生根增长,不定根和侧根密度降低 [46] OsLOGL5 Os03g0857900 OsLOGL5表达量的改变影响根系伸长和侧根发育 [47] BG3 Os01g0680200 bg3-D根生长量减少 [48] OsBRD1 Os03g0602300 osbrd1根系发育不良,冠根数量少 [49] OsBRI1 Os01g0718300 osbri1根系生长异常 [50] OsBZR1 Os07g0580500 OsBZR1过表达植株侧根数增加 [49] OsKS1 Os04g0611800 ks1突变体根长较短和根分生组织更小 [51] OsSWEET3a Os05g0214300 OsSWEET3a的敲除系和过表达系均表现出根生长迟缓 [52] OsSLR1 Os03g0707600 突变体slr1根长较短且数目减少 [53] OsNCED2 Os12g0435200 干旱条件下,过表达植株冠根和侧根数增加,敲除突变体相反 [54] OsACS1 Os03g0727600 OsACS1过表达植株根系更短 [55] OsERS2 Os05g0155200 osers2突变体植株具有短根表型 [56] OsCOI2 Os03g0265500 MeJA处理后的coi2根比野生型长 [57] AIM1 Os02g0274100 突变体aim1根长和不定根根长显著变短 [58] -
[1] CHANG C R,HU Y B,SUN S B,et al. Proton pump OsA8 is linked to phosphorus uptake and translocation in rice[J]. Journal of Experimental Botany,2009,60(2) :557−565. DOI: 10.1093/jxb/ern298
[2] SUN Y F,LUO W Z,JAIN A,et al. OsPHR3 affects the traits governing nitrogen homeostasis in rice[J]. BMC Plant Biology,2018,18(1) :241. DOI: 10.1186/s12870-018-1462-7
[3] DAI X Y,WANG Y Y,ZHANG W H. OsWRKY74,a WRKY transcription factor,modulates tolerance to phosphate starvation in rice[J]. Journal of Experimental Botany,2016,67(3) :947−960. DOI: 10.1093/jxb/erv515
[4] LI K N,ZHANG S N,TANG S,et al. The rice transcription factor Nhd1 regulates root growth and nitrogen uptake by activating nitrogen transporters[J]. Plant Physiology,2022,189(3) :1608−1624. DOI: 10.1093/plphys/kiac178
[5] EL-KEREAMY A,BI Y M,MAHMOOD K,et al. Overexpression of the CC-type glutaredoxin,OsGRX6 affects hormone and nitrogen status in rice plants[J]. Frontiers in Plant Science,2015,6:934.
[6] CAO Y,YAN Y,ZHANG F,et al. Fine characterization of OsPHO2 knockout mutants reveals its key role in Pi utilization in rice[J]. Journal of Plant Physiology,2014,171(3/4) :340−348.
[7] ZHOU S L,JIANG W,LONG F,et al. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem[J]. The Plant Cell,2017,29(5) :1088−1104. DOI: 10.1105/tpc.16.00908
[8] SHI M,XIE Y R,ZHENG Y Y,et al. Oryza sativa actin-interacting protein 1 is required for rice growth by promoting actin turnover[J]. Plant Journal,2013,73(5) :747−760. DOI: 10.1111/tpj.12065
[9] XU L,WANG F,LI R L,et al. OsCYCP4s coordinate phosphate starvation signaling with cell cycle progression in rice[J]. Journal of Integrative Plant Biology,2020,62(7) :1017−1033. DOI: 10.1111/jipb.12885
[10] KIM C M,PARK S H,JE B I,et al. OsCSLD1,a cellulose synthase-like D1 gene,is required for root hair morphogenesis in rice[J]. Plant Physiology,2007,143(3) :1220−1230. DOI: 10.1104/pp.106.091546
[11] INUKAI Y,SAKAMOTO T,MORINAKA Y,et al. ROOT GROWTH INHIBITING,a rice endo-1,4-β-d-glucanase,regulates cell wall loosening and is essential for root elongation[J]. Journal of Plant Growth Regulation,2012,31(3) :373−381. DOI: 10.1007/s00344-011-9247-3
[12] XIONG G Y,LI R,QIAN Q,et al. The rice dynamin-related protein DRP2B mediates membrane trafficking,and thereby plays a critical role in secondary cell wall cellulose biosynthesis[J]. Plant Journal,2010,64(1) :56−70.
[13] QU L H,WU C Y,ZHANG F,et al. Rice putative methyltransferase gene OsTSD2 is required for root development involving pectin modification[J]. Journal of Experimental Botany,2016,67(18) :5349−5362. DOI: 10.1093/jxb/erw297
[14] PARK J J,YI J,YOON J,et al. OsPUB15,an E3 ubiquitin ligase,functions to reduce cellular oxidative stress during seedling establishment[J]. Plant Journal,2011,65(2) :194−205. DOI: 10.1111/j.1365-313X.2010.04416.x
[15] JOO J,LEE Y H,SONG S I. Rice CatA,CatB,and CatC are involved in environmental stress response,root growth,and photorespiration,respectively[J]. Journal of Plant Biology,2014,57(6) :375−382. DOI: 10.1007/s12374-014-0383-8
[16] LU Y Z,FENG Z,LIU X Y,et al. MiR393 and miR390 synergistically regulate lateral root growth in rice under different conditions[J]. BMC Plant Biology,2018,18(1) :261. DOI: 10.1186/s12870-018-1488-x
[17] MORITA S,NEMOTO K. Morphology and anatomy of rice roots with special reference to coordination in organo- and histogenesis[M]//Structure and Function of Roots. Dordrecht:Springer Netherlands,1995:75–86.
[18] SUN H W,LI W Q,BURRITT D J,et al. Strigolactones interact with other phytohormones to modulate plant root growth and development[J]. The Crop Journal,2022,10(6) :1517−1527. DOI: 10.1016/j.cj.2022.07.014
[19] ROBBINS N E II,DINNENY J R. Growth is required for perception of water availability to pattern root branches in plants[J]. Proceedings of the National Academy of Sciences of the United States of America,2018,115(4) :E822−E831.
[20] SEBASTIAN J,YEE M C,GOUDINHO VIANA W,et al. Grasses suppress shoot-borne roots to conserve water during drought[J]. Proceedings of the National Academy of Sciences of the United States of America,2016,113(31) :8861−8866.
[21] BENGOUGH A G,LOADES K,MCKENZIE B M. Root hairs aid soil penetration by anchoring the root surface to pore walls[J]. Journal of Experimental Botany,2016,67(4) :1071−1078. DOI: 10.1093/jxb/erv560
[22] CLARK L H,HARRIS W H. Observations on the root anatomy of rice (Oryza sativa L. ) [J]. American Journal of Botany,1981,68(2) :154. DOI: 10.1002/j.1537-2197.1981.tb12374.x
[23] MOTTE H,VANNESTE S,BEECKMAN T. Molecular and environmental regulation of root development[J]. Annual Review of Plant Biology,2019,70:465−488. DOI: 10.1146/annurev-arplant-050718-100423
[24] CARRILLO-CARRASCO V P,HERNANDEZ-GARCIA J,MUTTE S K,et al. The birth of a giant:Evolutionary insights into the origin of auxin responses in plants[J]. The EMBO Journal,2023,42(6) :e113018. DOI: 10.15252/embj.2022113018
[25] YAMAZAKI K,FUJIWARA T. The effect of phosphate on the activity and sensitivity of nutritropism toward ammonium in rice roots[J]. Plants,2022,11(6) :733. DOI: 10.3390/plants11060733
[26] WANG H Q,ZHAO X Y,XUAN W,et al. Rice roots avoid asymmetric heavy metal and salinity stress via an RBOH-ROS-auxin signaling cascade[J]. Molecular Plant,2023,16(10) :1678−1694. DOI: 10.1016/j.molp.2023.09.007
[27] ZHU S X,ZHAO W,SUN S X,et al. Community metagenomics reveals the processes of cadmium resistance regulated by microbial functions in soils with Oryza sativa root exudate input[J]. Science of the Total Environment,2024,949:175015. DOI: 10.1016/j.scitotenv.2024.175015
[28] WU B B,WANG J Y,DAI H Y,et al. Radial oxygen loss triggers diel fluctuation of cadmium dissolution in the rhizosphere of rice[J]. Environmental Science & Technology,2024,58(33) :14718−14725.
[29] HUANG Y Z,JI Z,TAO Y J,et al. Improving rice nitrogen-use efficiency by modulating a novel monouniquitination machinery for optimal root plasticity response to nitrogen[J]. Nature Plants,2023,9(11) :1902−1914. DOI: 10.1038/s41477-023-01533-7
[30] LEI Z L,DING Y X,XU W F,et al. Microbial community structure in rice rhizosheaths under drought stress[J]. Journal of Plant Ecology,2023,16(5) :rtad012. DOI: 10.1093/jpe/rtad012
[31] DING Z J,XU C,YAN J Y,et al. The LRR receptor-like kinase ALR1 is a plant aluminum ion sensor[J]. Cell Research,2024,34(4) :281−294. DOI: 10.1038/s41422-023-00915-y
[32] RONZAN M,PIACENTINI D,FATTORINI L,et al. Cadmium and arsenic affect root development in Oryza sativa L. negatively interacting with auxin[J]. Environmental and Experimental Botany,2018,151:64−75. DOI: 10.1016/j.envexpbot.2018.04.008
[33] QIN H,HUANG R F. The phytohormonal regulation of Na+/K+ and reactive oxygen species homeostasis in rice salt response[J]. Molecular Breeding,2020,40(5) :47. DOI: 10.1007/s11032-020-1100-6
[34] ARSOVA B,FOSTER K J,SHELDEN M C,et al. Dynamics in plant roots and shoots minimize stress,save energy and maintain water and nutrient uptake[J]. New Phytologist,2020,225(3) :1111−1119. DOI: 10.1111/nph.15955
[35] JIA Z T,GIEHL R F H,VON WIRÉN N. Local auxin biosynthesis acts downstream of brassinosteroids to trigger root foraging for nitrogen[J]. Nature Communications,2021,12(1) :5437. DOI: 10.1038/s41467-021-25250-x
[36] LI J H,ZHANG Z Y,CHONG K,et al. Chilling tolerance in rice:Past and present[J]. Journal of Plant Physiology,2022,268:153576. DOI: 10.1016/j.jplph.2021.153576
[37] GROVER M,BODHANKAR S,SHARMA A,et al. PGPR mediated alterations in root traits:Way toward sustainable crop production[J]. Frontiers in Sustainable Food Systems,2021,4:618230. DOI: 10.3389/fsufs.2020.618230
[38] WU Q Q,PENG X J,YANG M F,et al. Rhizobia promote the growth of rice shoots by targeting cell signaling,division and expansion[J]. Plant Molecular Biology,2018,97(6) :507−523. DOI: 10.1007/s11103-018-0756-3
[39] XU F Y,LIAO H P,ZHANG Y J,et al. Coordination of root auxin with the fungus Piriformospora indica and bacterium Bacillus cereus enhances rice rhizosheath formation under soil drying[J]. The ISME Journal,2022,16(3) :801−811. DOI: 10.1038/s41396-021-01133-3
[40] MEENA K K,BITLA U M,SORTY A M,et al. Mitigation of salinity stress in wheat seedlings due to the application of phytohormone-rich culture filtrate extract of methylotrophic actinobacterium Nocardioides sp. NIMMe6[J]. Frontiers in Microbiology,2020,11:2091.
[41] ZHANG Y J,DU H,XU F Y,et al. Root-bacteria associations boost rhizosheath formation in moderately dry soil through ethylene responses[J]. Plant Physiology,2020,183(2) :780−792. DOI: 10.1104/pp.19.01020
[42] ZHANG T,LI R N,XING J L,et al. The YUCCA-auxin-WOX11 module controls crown root development in rice[J]. Frontiers in Plant Science,2018,9:523. DOI: 10.3389/fpls.2018.00523
[43] YOSHIKAWA T,ITO M,SUMIKURA T,et al. The rice FISH BONE gene encodes a tryptophan aminotransferase,which affects pleiotropic auxin-related processes[J]. Plant Journal,2014,78(6) :927−936. DOI: 10.1111/tpj.12517
[44] SUN C D,LI D M,GAO Z Y,et al. OsRLR4 binds to the OsAUX1 promoter to negatively regulate primary root development in rice[J]. Journal of Integrative Plant Biology,2022,64(1) :118−134. DOI: 10.1111/jipb.13183
[45] LI W Q,ZHANG M J,QIAO L,et al. Characterization of wavy root 1,an agravitropism allele,reveals the functions of OsPIN2 in fine regulation of auxin transport and distribution and in ABA biosynthesis and response in rice (Oryza sativa L. ) [J]. The Crop Journal,2022,10(4) :980−992. DOI: 10.1016/j.cj.2021.12.004
[46] GUO F,HUANG Y Z,QI P P,et al. Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield,tillering,plant height,root system,germination,and auxinic herbicide resistance[J]. New Phytologist,2021,229(5) :2676−2692. DOI: 10.1111/nph.17061
[47] CHEN L,JAMESON G B,GUO Y C,et al. The LONELY GUY gene family:From mosses to wheat,the key to the formation of active cytokinins in plants[J]. Plant Biotechnology Journal,2022,20(4) :625−645. DOI: 10.1111/pbi.13783
[48] XIAO Y H,LIU D P,ZHANG G X,et al. Big Grain3,encoding a purine permease,regulates grain size via modulating cytokinin transport in rice[J]. Journal of Integrative Plant Biology,2019,61(5) :581−597. DOI: 10.1111/jipb.12727
[49] JIAO X M,WANG H C,YAN J J,et al. Promotion of BR biosynthesis by miR444 is required for ammonium-triggered inhibition of root growth[J]. Plant Physiology,2020,182(3) :1454−1466. DOI: 10.1104/pp.19.00190
[50] 赵雪松,王倩,闫青地,等. 油菜素内酯对水稻根系发育的调控作用[J]. 中国细胞生物学学报,2016,38(10) :1191−1198. DOI: 10.11844/cjcb.2016.10.0109 ZHAO X S,WANG Q,YAN Q D,et al. Function of brassinolide in the regulation of root development in rice[J]. Chinese Journal of Cell Biology,2016,38(10) :1191−1198. (in Chinese) DOI: 10.11844/cjcb.2016.10.0109
[51] HE Y Q,HONG G J,ZHANG H H,et al. The OsGSK2 kinase integrates brassinosteroid and jasmonic acid signaling by interacting with OsJAZ4[J]. The Plant Cell,2020,32(9) :2806−2822. DOI: 10.1105/tpc.19.00499
[52] MORII M,SUGIHARA A,TAKEHARA S,et al. The dual function of OsSWEET3a as a gibberellin and glucose transporter is important for young shoot development in rice[J]. Plant & Cell Physiology,2020,61(11) :1935−1945.
[53] DAVIÈRE J M,ACHARD P. A pivotal role of DELLAs in regulating multiple hormone signals[J]. Molecular Plant,2016,9(1) :10−20. DOI: 10.1016/j.molp.2015.09.011
[54] HUANG L Y,BAO Y C,QIN S W,et al. The ABA synthesis enzyme allele OsNCED2T promotes dryland adaptation in upland rice[J]. The Crop Journal,2024,12(1) :68−78. DOI: 10.1016/j.cj.2023.12.001
[55] QIN H,WANG J,CHEN X B,et al. Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress[J]. New Phytologist,2019,223(2) :798−813. DOI: 10.1111/nph.15824
[56] ZHAO H,DUAN K X,MA B,et al. Histidine kinase MHZ1/OsHK1 interacts with ethylene receptors to regulate root growth in rice[J]. Nature Communications,2020,11(1) :518. DOI: 10.1038/s41467-020-14313-0
[57] INAGAKI H,HAYASHI K,TAKAOKA Y,et al. Genome editing reveals both the crucial role of OsCOI2 in jasmonate signaling and the functional diversity of COI1 homologs in rice[J]. Plant & Cell Physiology,2023,64(4) :405−421.
[58] XU L,ZHAO H Y,RUAN W Y,et al. ABNORMAL INFLORESCENCE MERISTEM1 functions in salicylic acid biosynthesis to maintain proper reactive oxygen species levels for root meristem activity in rice[J]. The Plant Cell,2017,29(3) :560−574. DOI: 10.1105/tpc.16.00665
[59] WAADT R,SELLER C A,HSU P K,et al. Plant hormone regulation of abiotic stress responses[J]. Nature Reviews Molecular Cell Biology,2022,23(10) :680−694. DOI: 10.1038/s41580-022-00479-6
[60] ZHAO B Q,LIU Q Y,WANG B S,et al. Roles of phytohormones and their signaling pathways in leaf development and stress responses[J]. Journal of Agricultural and Food Chemistry,2021,69(12) :3566−3584. DOI: 10.1021/acs.jafc.0c07908
[61] MAO C J,HE J M,LIU L N,et al. OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development[J]. Plant Biotechnology Journal,2020,18(2) :429−442. DOI: 10.1111/pbi.13209
[62] ZHAO J,YANG B,LI W J,et al. A genome-wide association study reveals that the glucosyltransferase OsIAGLU regulates root growth in rice[J]. Journal of Experimental Botany,2021,72(4) :1119−1134. DOI: 10.1093/jxb/eraa512
[63] ZHANG S Z,WU T,LIU S J,et al. Disruption of OsARF19 is critical for floral organ development and plant architecture in rice (Oryza sativa L. ) [J]. Plant Molecular Biology Reporter,2016,34(4) :748−760. DOI: 10.1007/s11105-015-0962-y
[64] WANG M,QIAO J Y,YU C L,et al. The auxin influx carrier,OsAUX3,regulates rice root development and responses to aluminium stress[J]. Plant,Cell & Environment,2019,42(4) :1125–1138.
[65] YE R G,WU Y R,GAO Z Y,et al. Primary root and root hair development regulation by OsAUX4 and its participation in the phosphate starvation response[J]. Journal of Integrative Plant Biology,2021,63(8) :1555−1567. DOI: 10.1111/jipb.13142
[66] JIANG L H,YAO B L,ZHANG X Y,et al. Salicylic acid inhibits rice endocytic protein trafficking mediated by OsPIN3t and clathrin to affect root growth[J]. Plant Journal,2023,115(1) :155−174. DOI: 10.1111/tpj.16218
[67] GAO J,ZHAO Y,ZHAO Z K,et al. RRS1 shapes robust root system to enhance drought resistance in rice[J]. New Phytologist,2023,238(3) :1146−1162. DOI: 10.1111/nph.18775
[68] CHEN Y,YANG Q F,SANG S H,et al. Rice inositol polyphosphate kinase (OsIPK2) directly interacts with OsIAA11 to regulate lateral root formation[J]. Plant & Cell Physiology,2017,58(11) :1891−1900.
[69] KITOMI Y,INAHASHI H,TAKEHISA H,et al. OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice[J]. Plant Science,2012,190:116−122. DOI: 10.1016/j.plantsci.2012.04.005
[70] NI J,WANG G H,ZHU Z X,et al. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice[J]. Plant Journal,2011,68(3) :433−442. DOI: 10.1111/j.1365-313X.2011.04698.x
[71] QI Y H,WANG S K,SHEN C J,et al. OsARF12,a transcription activator on auxin response gene,regulates root elongation and affects iron accumulation in rice (Oryza sativa) [J]. New Phytologist,2012,193(1) :109−120. DOI: 10.1111/j.1469-8137.2011.03910.x
[72] WANG X F,HE F F,MA X X,et al. OsCAND1 is required for crown root emergence in rice[J]. Molecular Plant,2011,4(2) :289−299. DOI: 10.1093/mp/ssq068
[73] HAN Y F,ZHANG C Z,SHA H J,et al. Ubiquitin-conjugating enzyme OsUBC11 affects the development of roots via auxin pathway[J]. Rice,2023,16(1) :9. DOI: 10.1186/s12284-023-00626-3
[74] WYBOUW B,DE RYBEL B. Cytokinin - A developing story[J]. Trends in Plant Science,2019,24(2) :177−185. DOI: 10.1016/j.tplants.2018.10.012
[75] ZHAO J Z,YU N N,JU M,et al. ABC transporter OsABCG18 controls the shootward transport of cytokinins and grain yield in rice[J]. Journal of Experimental Botany,2019,70(21) :6277−6291. DOI: 10.1093/jxb/erz382
[76] GAO S P,FANG J,XU F,et al. CYTOKININ OXIDASE/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation[J]. Plant Physiology,2014,165(3) :1035−1046. DOI: 10.1104/pp.114.238584
[77] DO NASCIMENTO F C,DE SOUZA A F F,DE SOUZA V M,et al. OsCKX5 modulates root system morphology and increases nutrient uptake in rice[J]. Journal of Plant Growth Regulation,2022,41(6) :2157−2170. DOI: 10.1007/s00344-021-10419-x
[78] NONGPIUR R C,RAWAT N,SINGLA-PAREEK S L,et al. OsRR26,a type-B response regulator,modulates salinity tolerance in rice via phytohormone-mediated ROS accumulation in roots and influencing reproductive development[J]. Planta,2024,259(5) :96. DOI: 10.1007/s00425-024-04366-6
[79] LIU H L,HUANG J Q,ZHANG X J,et al. The RAC/ROP GTPase activator OsRopGEF10 functions in crown root development by regulating cytokinin signaling in rice[J]. The Plant Cell,2023,35(1) :453−468. DOI: 10.1093/plcell/koac297
[80] HOU J Q,ZHENG X K,REN R F,et al. The histone deacetylase 1/GSK3/SHAGGY-like kinase 2/BRASSINAZOLE-RESISTANT 1 module controls lateral root formation in rice[J]. Plant Physiology,2022,189(2) :858−873. DOI: 10.1093/plphys/kiac015
[81] JIANG Y H,BAO L,JEONG S Y,et al. XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice[J]. Plant Journal,2012,70(3) :398−408. DOI: 10.1111/j.1365-313X.2011.04877.x
[82] LO S F,YANG S Y,CHEN K T,et al. A novel class of gibberellin 2-oxidases control semidwarfism,tillering,and root development in rice[J]. The Plant Cell,2008,20(10) :2603−2618. DOI: 10.1105/tpc.108.060913
[83] LI J T,ZHAO Y,CHU H W,et al. SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation[J]. PLoS Genetics,2015,11(8) :e1005464. DOI: 10.1371/journal.pgen.1005464
[84] MO W P,TANG W J,DU Y X,et al. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 interaction controls seedling growth under salt stress[J]. Plant Physiology,2020,184(1) :506−517. DOI: 10.1104/pp.20.00024
[85] LI J N,ZHANG Y X,LI Z Y,et al. OsPEX1,an extensin-like protein,negatively regulates root growth in a gibberellin-mediated manner in rice[J]. Plant Molecular Biology,2023,112(1/2) :47−59.
[86] TENG Z N,LYU J H,CHEN Y K,et al. Effects of stress-induced ABA on root architecture development:Positive and negative actions[J]. The Crop Journal,2023,11(4) :1072−1079. DOI: 10.1016/j.cj.2023.06.007
[87] CHEN H,MA B,ZHOU Y,et al. E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein[J]. Proceedings of the National Academy of Sciences of the United States of America,2018,115(17) :4513−4518.
[88] SANTOSH KUMAR V V,YADAV S K,VERMA R K,et al. The abscisic acid receptor OsPYL6 confers drought tolerance to indica rice through dehydration avoidance and tolerance mechanisms[J]. Journal of Experimental Botany,2021,72(4) :1411−1431. DOI: 10.1093/jxb/eraa509
[89] XU N,CHU Y L,CHEN H L,et al. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging[J]. PLoS Genetics,2018,14(10) :e1007662. DOI: 10.1371/journal.pgen.1007662
[90] KAWAI T,SHIBATA K,AKAHOSHI R,et al. WUSCHEL-related homeobox family genes in rice control lateral root primordium size[J]. Proceedings of the National Academy of Sciences of the United States of America,2022,119(1) :e2101846119.
[91] YOON J,CHO L H,YANG W Z,et al. Homeobox transcription factor OsZHD2 promotes root meristem activity in rice by inducing ethylene biosynthesis[J]. Journal of Experimental Botany,2020,71(18) :5348−5364. DOI: 10.1093/jxb/eraa209
[92] MA B,YIN C C,HE S J,et al. Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L. ) seedlings[J]. PLoS Genetics,2014,10(10) :e1004701. DOI: 10.1371/journal.pgen.1004701
[93] ZHAO H,MA B,DUAN K X,et al. The GDSL lipase MHZ11 modulates ethylene signaling in rice roots[J]. The Plant Cell,2020,32(5) :1626−1643. DOI: 10.1105/tpc.19.00840
[94] MA B,ZHOU Y,CHEN H,et al. Membrane protein MHZ3 stabilizes OsEIN2 in rice by interacting with its Nramp-like domain[J]. Proceedings of the National Academy of Sciences of the United States of America,2018,115(10) :2520−2525.
[95] ZHOU Y,GAO Y H,ZHANG B C,et al. CELLULOSE SYNTHASE-LIKE C proteins modulate cell wall establishment during ethylene-mediated root growth inhibition in rice[J]. The Plant Cell,2024,36(9) :3751−3769. DOI: 10.1093/plcell/koae195
[96] WU F H,GAO Y P,YANG W J,et al. Biological functions of strigolactones and their crosstalk with other phytohormones[J]. Frontiers in Plant Science,2022,13:821563. DOI: 10.3389/fpls.2022.821563
[97] WANG B B,ZHU X L,GUO X L,et al. Nitrate modulates lateral root formation by regulating the auxin response and transport in rice[J]. Genes,2021,12(6) :850. DOI: 10.3390/genes12060850
[98] SUN H W,GUO X L,QI X J,et al. SPL14/17 act downstream of strigolactone signalling to modulate rice root elongation in response to nitrate supply[J]. Plant Journal,2021,106(3) :649−660. DOI: 10.1111/tpj.15188
[99] RAYA-GONZÁLEZ J,ORTIZ-CASTRO R,RUÍZ-HERRERA L F,et al. PHYTOCHROME AND FLOWERING TIME1/MEDIATOR25 regulates lateral root formation via auxin signaling in Arabidopsis[J]. Plant Physiology,2014,165(2) :880−894. DOI: 10.1104/pp.114.239806
[100] SUZUKI G,LUCOB-AGUSTIN N,KASHIHARA K,et al. Rice MEDIATOR25,OsMED25,is an essential subunit for jasmonate-mediated root development and OsMYC2-mediated leaf senescence[J]. Plant Science,2021,306:110853. DOI: 10.1016/j.plantsci.2021.110853
[101] MUZAFFAR A,CHEN Y S,LEE H T,et al. A newly evolved rice-specific gene JAUP1 regulates jasmonate biosynthesis and signalling to promote root development and multi-stress tolerance[J]. Plant Biotechnology Journal,2024,22(5) :1417−1432. DOI: 10.1111/pbi.14276
[102] WATERS M T,GUTJAHR C,BENNETT T,et al. Strigolactone signaling and evolution[J]. Annual Review of Plant Biology,2017,68:291−322. DOI: 10.1146/annurev-arplant-042916-040925
[103] XIANG D,MENG F N,WANG A D,et al. Root-secreted peptide OsPEP1 regulates primary root elongation in rice[J]. Plant Journal,2021,107(2) :480−492. DOI: 10.1111/tpj.15303
[104] LI J Y,MENG L J,REN S H,et al. OsGSTU17,a tau class glutathione S-transferase gene,positively regulates drought stress tolerance in Oryza sativa[J]. Plants,2023,12(17) :3166. DOI: 10.3390/plants12173166
[105] YANG H S,FANG Y Y,LIANG Z M,et al. Polyamines:Pleiotropic molecules regulating plant development and enhancing crop yield and quality[J]. Plant Biotechnology Journal,2024,22(11) :3194−3201. DOI: 10.1111/pbi.14440
[106] MAI H F,QIN T,WEI H,et al. Overexpression of OsACL5 triggers environmentally-dependent leaf rolling and reduces grain size in rice[J]. Plant Biotechnology Journal,2024,22(4) :833−847. DOI: 10.1111/pbi.14227
[107] GAO Y Q,GUO R,WANG H Y,et al. Melatonin increases root cell wall phosphorus reutilization via an NO dependent pathway in rice (Oryza sativa) [J]. Journal of Pineal Research,2024,76(5) :e12995. DOI: 10.1111/jpi.12995




 下载:
下载: