Characteristics and Evolution of Panax Chloroplast Genomes
-
摘要:目的 对人参属(Panax L.)物种叶绿体基因组特征及其系统发育进行研究,为我国人参属资源的遗传学研究和开发利用提供理论依据。方法 基于14种人参属植物叶绿体基因组序列,利用生物信息学软件,对叶绿体基因组特征、序列重复、结构变异、基因进化和系统发育进行分析。结果 人参属物种叶绿体基因组均为典型的四分体结构,包含114个unique基因。长重复序列主要为回文重复和正向重复,30~39 bp的重复序列最多,SSR大多为A/T重复,单核苷酸重复是最丰富的类型。人参属叶绿体基因组未发生基因重排,反向重复区(IR)与单拷贝区边界高度保守,鉴定的12个核苷酸高度可变热点中7个位于大单拷贝(LSC)区,5个位于小单拷贝(SSC)区。根据dN/dS比率,发现功能未知基因clpP、ycf1和ycf2受正选择作用。系统发育分析显示,屏边三七和三叶参位于基部支系,四倍体人参和西洋参与其他二倍体物种聚在不同支系,三七、竹节参和越南参则亲缘关系较近。结论 人参属叶绿体基因组基因数目和顺序一致,基因组结构保守,重复序列数目和类型存在差异,单拷贝区核苷酸多态性高于IR区,正选择基因可能与物种的生态适应性有关。Abstract:Objective Characteristics and phylogeny of chloroplast genomes of the medicinally and economically valuable species in Panax genus were studied.Methods Using bioinformatics software, the properties, repeats, structural variation, evolution, and phylogeny of the genomes of chloroplasts from 14 ginseng species were analyzed.Results The genomes consisted of typical quadripartite structure with 114 unique genes. The long repeats in them were mainly of palindromic and forward types with a length between 30 bp and 39 bp. The simple sequence repeats were largely A/T type and most abundantly mononucleotides. No gene rearrangement occurred in the genomes was observed. The boundary between the inverted repeat region and the single copy region was highly conserved. Of the 12 regions with highly variable nucleotides, 7 were in the large and 5 in the small single copy region. Indicated by the dN/dS ratios, the positive selection could occur on clpP, ycf1, and ycf2 with unknown functions. The phylogenetic analysis showed that P. stipulenatus and P. trifolius were in the basal lineage, the tetraploid P. ginseng and P. quinquefolius separated from other diploid species, while P. notoginseng, P. japonicus, and P. vietnamensis closely related.Conclusion The chloroplasts of the ginseng species examined were basically same in number and order of genomes, conservative in structure, but divert in number and type of repeats. The nucleotide polymorphism of the chloroplasts was higher in single copy region than inverted repeat regions. The positive selection genes identified in the study might result from the ecological adaptation of these Panax species.
-
Keywords:
- Panax L. /
- chloroplast genome /
- repeat /
- variation /
- phylogeny
-
0. 引言
【研究意义】人参属(Panax L.)属伞形目五加科(Araliaceae)多年生草本植物,起源于喜马拉雅山区[1],主要分布于东亚和北美的高海拔山区,该属几乎所有物种均具有重要的药用价值,特别根茎被广泛用作草药[2]。叶绿体是绿色植物体重要的细胞器,植物物种和个体之间的叶绿体基因组变异,对于植物群体遗传学研究和物种鉴定均具有重要价值。叶绿体基因组研究可为人参属物种鉴定、起源、进化、遗传多样性分析和资源保护与利用提供基础。【前人研究进展】人参属包含14个物种,其中人参(P. ginseng)、西洋参(P. quinquefolius)、三七(P. notoginseng)、竹节参(P. japonicus)和越南参(P. vietnamensis)为中国、美国、日本和越南等地贵细类药材,用于治疗和预防多种疾病,具有很大国际市场[3]。人参、屏边三七(P. stipuleanatus)和姜状三七(P. zingiberensis)均已入选中国珍稀濒危植物名录,其中人参属植物具有相似的形态特征,但在活性成分和药理作用上存在明显差异[4-5]。人参富含100余种人参皂苷类活性成分,是我国中草药材中著名的植物类药材,有助于缓解疲劳、促进康复和预防潜在疾病[5],其富含的人参皂苷Ra2、Rg1、Ra1以及丙二酰人参皂苷Ra3等,可通过作用于哺乳动物雷帕霉素靶蛋白(mTOR)、胞内磷脂酰肌醇激酶(PI3K)和其他靶点来调节前列腺特异性抗原(PSA)、核糖体S6激酶(S6K)、MDM2(Murine double minute 2)和P53基因的mRNA表达,从而抑制乳腺癌的增殖[4]。西洋参产自美国和加拿大南部,主要用于治疗气虚性贫血、咳嗽和哮喘[6]。三七主要分布在中国云南省,含有皂苷和三七素等止血活性成分,常用于调节和治疗心血管系统、神经系统和免疫系统等方面疾病[7]。叶绿体基因组编码多个参与光合作用等重要代谢反应的关键蛋白,由一对反向重复区(Inverted repeats,IRs)分别将大单拷贝区(Large single copy,LSC)和小单拷贝区(Small single copy,SSC)分开,从而构成典型的四分体结构[2,8-11]。大部分被子植物叶绿体基因组包含120~130个基因,其中包括编码光合作用相关的蛋白编码基因、核糖体RNA基因和转运RNA基因[8,11]。随着高通量测序技术的快速发展,大量植物叶绿体基因组序列已完成测序,叶绿体基因组被广泛用于物种鉴定和系统发育分析[8,11-12]。【本研究切入点】目前,多个人参属物种叶绿体基因组序列已被公布[13-14],鉴于人参属物种重要的开发和利用前景,应用叶绿体基因组的遗传分析,对于人参属物种鉴定、资源保护和开发利用均具有重要的理论和实践意义。【拟解决的关键问题】本研究基于已公布的叶绿体基因组数据,利用生物信息学方法,对人参属物种叶绿体基因组结构、重复序列、核苷酸变异位点、基因进化以及系统发育进行分析。人参属物种亲缘关系和遗传多样性分析,将为我国人参属资源的品种改良和开发利用提供理论依据。
1. 材料与方法
1.1 数据收集
从美国国家生物技术信息中心GenBank数据库(https://www.ncbi.nlm.nih.gov/)检索并下载14种人参属植物叶绿体基因组序列信息(表1)。
表 1 人参属物种叶绿体基因组特征Table 1. Chloroplast genomes of Panax species物种 Species 登录号 Accession 长度 Length/bp GC含量 GC content/% 基因组 Genome LSC SSC IR 基因组 Genome LSC SSC IR 人参
P. ginsengMK408938 156 241 86 128 18 077 26 018 38.07 36.27 32.18 43.10 竹节参
P. japonicusKP036469 156 188 86 199 18 013 25 988 38.07 36.29 32.21 43.05 疙瘩七
P. japonicus var. bipinnatifidusMK408962 156 244 86 186 18 006 26 026 38.06 36.28 32.21 43.05 珠子参
P. majorMN496312 156 402 86 189 18 007 26 103 38.07 36.28 32.22 43.05 三七
P. notoginsengMK408945 156 319 86 157 18 004 26 079 38.07 36.27 32.23 43.07 假人参
P. pseudoginsengMW145449 156 074 86 124 18 008 25 971 38.06 36.27 32.22 43.06 西洋参
P. quinquefoliusMK408953 156 070 86 077 17 993 26 000 38.07 36.27 32.22 43.08 屏边三七
P. stipuleanatusMK408965 156 007 86 083 18 150 25 887 38.04 36.26 32.07 43.08 三叶参
P. trifoliusMF100782 156 157 86 322 18 047 25 894 38.08 36.27 32.26 43.10 越南参
P. vietnamensisKP036470 155 993 86 178 17 935 25 940 38.05 36.27 32.22 43.02 野三七
P. vietnamensis var. fuscidiscusMT798585 156 284 86 171 17 971 26 071 38.06 36.28 32.27 42.98 越南参变种
P. vietnamensis var. langbianensisMT798583 155 984 86 174 17 934 25 938 38.05 36.27 32.21 43.03 峨眉三七
P. wangianusMK408964 156 189 86 190 17 969 26 015 38.07 36.29 32.21 43.03 姜状三七
P. zingiberensisMK408969 156 192 86 116 17 966 26 055 38.07 36.31 32.27 43.00 1.2 试验方法
1.2.1 重复序列分析
通过REPuter软件[15]分析长重复序列,参数设置为最小重复长度30 bp,最大碱基错配数3。通过MISA软件[16]检测简单重复序列(Simple sequence repeat,SSR)数目及位置分布,最小重复数设为单核苷酸10,二核苷酸5,三核苷酸4,四核苷酸、五核苷酸和六核苷酸均为3。
1.2.2 基因组结构分析
使用IRscope软件[17]对人参属物种叶绿体基因组LSC、SSC和IRs边界的收缩与扩张进行可视化。通过mVISTA软件[18],以人参叶绿体基因组序列(MK408938)为参照,选用Shuffle-LAGAN全局比对模式,对人参属物种叶绿体基因组序列同源性进行可视化比对。
1.2.3 核苷酸多样性分析
使用MAFFT软件[19]对叶绿体基因组序列进行比对,使用DnaSP软件[20]计算叶绿体基因组序列核苷酸多态性(Pi),参数设置为搜索窗口长度600 bp,步长200 bp,使用R程序绘图。
1.2.4 基因进化分析
使用MAFFT软件对各物种叶绿体蛋白编码基因进行比对,使用PAMLX[21]的CODEML算法采用位点模型法对正选择位点进行检测,并评价人参属物种叶绿体蛋白编码基因的进化情况。
1.2.5 系统发育分析
利用14种人参属植物叶绿体基因组序列,以五加科物种楤木(Aralia elata)和波缘楤木(A. undulata)为外类群,使用IQ-TREE 2软件[22]基于最大似然法(Maximum likelihood,ML)构建系统发育树,建树模型为TVM+F+R6,步长值为1 000。
2. 结果与分析
2.1 叶绿体基因组特征
14种人参属植物叶绿体基因组均为双链环形结构,由LSC、SSC、IRa和IRb等4部分构成,基因组总长度为155 984 bp(越南参变种P. vietnamensis var. langbianensis)~156 402 bp(珠子参P. major),属内叶绿体基因组长度差异小于500 bp,总GC含量为38.04%~38.08%,LSC、SSC和IR长度分别为86 077 ~86 322 bp、17 934 ~18 150 bp和25 887 ~26 103 bp,GC含量分别为36.26%~36.31%、32.07%~32.27%和42.98%~43.10%(表1)。所有基因组均包含114个unique基因,其中有80个蛋白编码基因,4个rRNA和30个tRNA。21个基因属于双拷贝基因,包括8个蛋白编码基因(ndhB、rpl2、rpl23、rps7、rps12、ycf1、ycf2和 ycf15),4个rRNA基因(rrn4.5、rrn5、rrn16和rrn23),7个tRNA基因(trnA-UGC、trnI-CAU、trnI-GAU、trnL-CAA、trnN-GUU、trnR-ACG和trnV-GAC)。3个基因(rps12、clpP 和 ycf3)包含2个内含子,15个基因包含1个内含子(atpF、ndhA、ndhB、petB、petD、rpl2、rpl16、rpoC1、rps16、trnA-UGC、trnG-UCC、trnI-GAU、trnK-UUU、trnL-UAA和trnV-UAC)。
2.2 重复序列分析
利用REPuter软件对14种人参属植物叶绿体基因组序列4种长重复序列进行分析,共检测到长度为30~100 bp的重复序列625条(图1)。4种重复类型中,回文重复数目最多,平均含有24条,其次为正向重复,平均20条,各物种中反向重复和互补重复数较少,除了野三七(P. vietnamensis var. fuscidiscus)中含有4条反向重复和2条互补重复外,其他物种仅含有0~1条反向重复,且均不含互补重复序列(图1-A)。根据重复序列长度分析显示,各物种中30~39 bp长度的序列最多,平均占56%,其次为40~49 bp序列,平均占23%,60~69 bp序列占15%,而50~59 bp和大于70 bp的序列占比均为3%(图1-B)。重复序列数目最多的是疙瘩七(P. japonicus var. bipinnatifidus)102条,最少的为屏边三七(P. stipuleanatus)27条,其他物种的重复序列数目为31~56条。
从14种人参属植物叶绿体基因组中共检测到574个SSR位点,各物种中分布数目为37~45个,单核苷酸重复数目最多,平均占49%,四核苷酸重复占21%,二核苷酸占15%,三核苷酸占7%,五核苷酸占6%,六核苷酸仅占2%(图2-A)。单核苷酸重复类型主要为A/T,占单碱基重复的83%;二核苷酸重复均为AT/AT型;三核苷酸重复有AAT/ATT、AAG/CTT和AGC/CTG等3种类型,其中AAT/ATT占60%;5种四核苷酸重复中,AAAG/CTTT型占42%,AAAT/ATTT型占36%,ACCT/AGGT型占28%,AATT/AATT型占14%,而AAAC/GTTT型仅占1%;五核苷酸重复主要为AATCT/AGATT型,占76%,部分物种中未检测到六核苷酸重复(图2-B)。人参属种间SSR数目差异相对较小,竹节参叶绿体基因组中SSR数目最多(45个),三七和疙瘩七的SSR最少(均为37个)。
2.3 叶绿体基因组结构变异
使用IRscope软件对人参属物种叶绿体基因组LSC/IRs/SSC边界收缩与扩张进行比较(图3)。14种植物IR区长度最大差异为216 bp,表明IR区存在一定程度的扩张或收缩。所有物种LSC/IRb边界均跨越基因rps19,IRb区延伸到rps19的长度为46~51 bp;IRb/SSC边界存在较大差异,7个物种IRb/SSC边界跨越基因ycf1,ycf1编码区延伸到SSC长度为1~3 bp,6个物种IRb/SSC边界跨越基因ndhF,IRb区延伸到ndhF长度为8~12 bp;所有物种SSC/IRa边界均位于基因ycf1编码区内,IRa区延伸到ycf1的长度为1 476~1 649 bp;全部物种IRa/LSC边界均跨越基因trnH,IRa的延伸长度为4~7 bp。
以人参叶绿体基因组(MK408938)为参照,使用mVISTA软件比较人参属物种叶绿体基因组序列,发现14种人参属植物叶绿体基因组序列一致性较高,非编码区序列变异高于编码区,LSC区和SSC区序列变异高于IR区。相比于人参属其他物种,屏边三七和三叶参序列变异相对较高,人参属叶绿体基因组基因数目和顺序一致,未发现基因重排现象(图4)。
2.4 叶绿体基因组核苷酸多态性
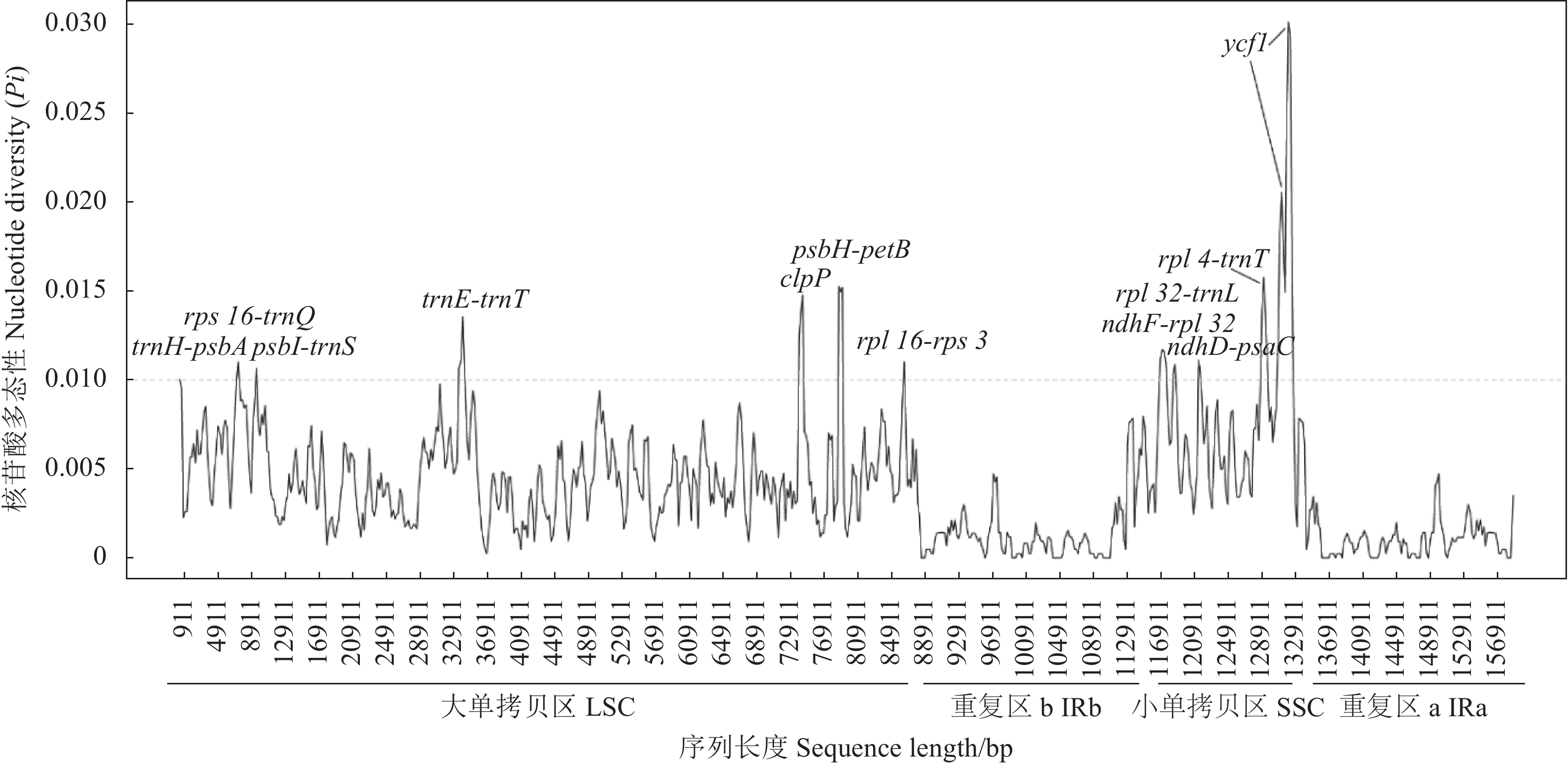
使用DnaSP软件对14种人参属植物叶绿体基因组序列核苷酸多态性(Pi)进行分析(图5),比对序列总长度为158 911 bp,共检测到多态性位点2 889个,核苷酸多样性值范围为0~0.030,平均值为0.004,LSC、SSC和IR区Pi平均值分别为0.005、0.008和0.001,显示LSC区和SSC区高度可变,IR区相对保守。鉴定的12个高度可变热点(Hotspot)中,7个位于LSC区(trnH-psbA、rps16-trnQ、psbI-trnS、trnE-trnT、clpP、psbH-petB和rpl16-rps3),5个位于SSC区(ndhF-rpl32、rpl32-trnL、ndhD-psaC、rps4-trnT和ycf1),IR区未检测到核苷酸多态性位点,表明单拷贝区核苷酸多态性明显高于IR区。
2.5 基因进化分析
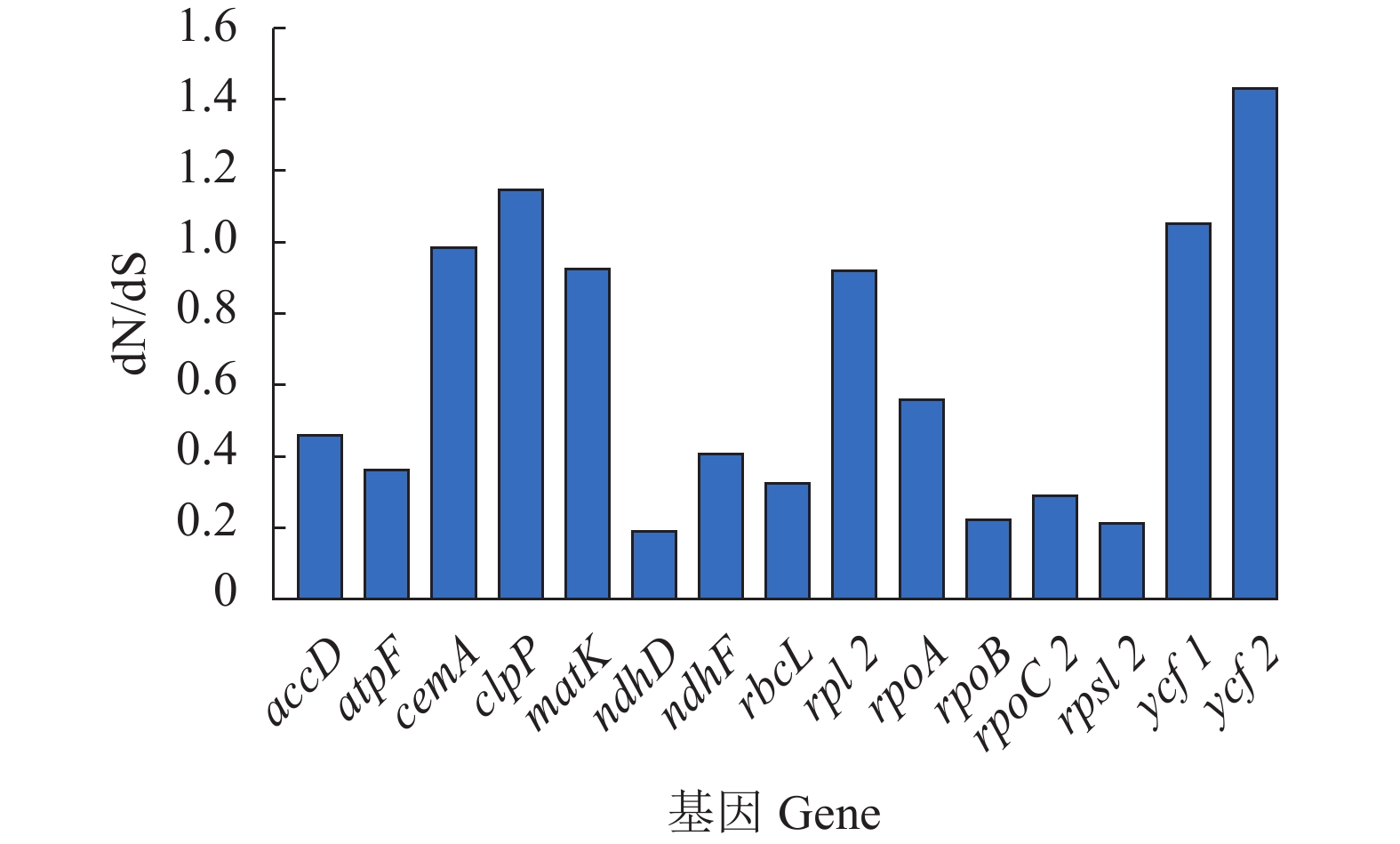
为了解人参属叶绿体蛋白编码基因在进化过程中是否受到选择作用,利用PAML软件对基因dN/dS比率和正选择位点进行分析(图6、表2)。结果发现,clpP、ycf1和ycf2的dN/dS比率均大于1,表明这些基因受到正选择作用。而cemA、matK和rpl2的dN/dS比率虽然小于1,但均大于0.92,表明这些基因进化过程中主要受纯化选择作用,同时也受到一定的选择作用影响。根据经验贝叶斯分析,共有8个蛋白编码基因正选择位点达显著水平(P>95%),分别为cemA、clpP、matK、ndhF、rbcL、rpoA、ycf1和ycf2,显著性正选择位点数分别为1、1、1、8、3、2、10和4(表2)。三叶参rpoA基因在803和804位插入两个碱基A造成移码突变,随后出现终止密码子TAG,导致蛋白翻译提前终止。
表 2 基于位点模型的人参属叶绿体蛋白编码基因正选择分析Table 2. Potential positive selection test on chloroplast genomes of Panax species based on site model基因
GeneM1 vs M2 M7 vs M8 正选择位点
Positively selected sites2ΔlnL P 值 2ΔlnL P 值 accD 2.39 3.0×10-1 2.47 2.9×10-1 139 Q, 0.505; 141 S, 0.517; 144 R, 0.528; 209 A, 0.856; 264 E, 0.504; 364 I, 0.507; 372 L, 0.526; 494 Q, 0.855 atpF 3.37 1.9×10-1 3.42 1.8×10-1 14 W, 0.541; 77 Q, 0.537; 158 G, 0.854 cemA 7.48 2.4×10-2 7.48 2.4×10-2 14 A, 0.547; 22 W, 0.538; 54 K, 0.536; 102 D, 0.533; 103 R, 0.544; 118 V, 0.973*; 205 F, 0.531; 208 W, 0.540 clpP 13.40 1.2×10-3 13.40 1.2×10-3 38 P, 0.990*; 39 V, 0.622; 40 A, 0.935; 41 S, 0.612; 138 G, 0.629; 160 Q, 0.626 matK 19.22 6.7×10-5 19.25 6.6×10-5 83 R, 0.994** ndhD 0.77 6.8×10-1 0.99 6.1×10-1 22 F, 0.830; 41 I, 0.829 ndhF 19.81 5.0×10-5 20.29 3.9×10-5 17 P, 0.597; 123 R, 0.592; 133 T, 0.583; 223 F, 0.959*; 421 I, 0.964*; 463 Q, 0.591; 483 R, 0.569; 499 H, 0.552; 502 A, 0.965*; 511 Q, 0.591; 512 M, 0.526; 532 K, 0.559; 534 I, 0.962*; 572 N, 0.543; 598 D, 0.573; 606 F, 0.548; 621 L, 0.630; 641 I, 0.549; 642 G, 0.597; 657 G, 0.594; 670 A, 0.963*; 677 I, 0.532; 703 S, 0.548; 737 Y, 0.964*; 743 F, 0.537; 745 D, 0.958*; 746 L, 1.000** rbcL 15.94 3.5×10-4 16.10 3.2×10-4 225 L, 0.632; 226 Y, 0.969*; 230 A, 0.635; 270 I, 0.866; 281 A, 0.644; 328 S, 0.999**; 353 F, 0.584; 439 R, 0.968*; 472 V, 0.609; 474 I, 0.621; 478 V, 0.714 rpl2 0 1 0 1 99 K, 0.651; 265 D, 0.650; 267 L, 0.652 rpoA 14.53 7.0×10-4 14.56 6.9×10-4 34 L, 0.506; 256 E, 0.991**; 264 N, 0.999**; 272 S, 0.533; 320 A, 0.849; 337 L, 0.537 rpoB 0.90 6.4×10-1 0.90 6.4×10-1 37 Y, 0.505; 165 R, 0.502; 193 R, 0.502; 295 I, 0.502; 365 T, 0.504; 459 S, 0.502; 695 K, 0.506; 735 T, 0.504; 935 D, 0.832 rpoC2 0 1 0 1 85 Q, 0.736 rps12 0 1 0.02 9.9×10-1 25 R, 0.502 ycf1 75.81 0 75.78 0 205 K, 1.000**; 206 Y, 0.999**; 288 I, 0.876; 532 T, 0.803; 533 K, 0.956*; 604 R, 0.801; 624 R, 0.825; 668 I, 0.779; 685 W, 0.837; 699 V, 0.824; 701 Q, 0.802; 706 V, 0.814; 720 V, 0.856; 722 T, 0.814; 723 D, 0.808; 731 R, 0.982*; 733 K, 0.809; 734 I, 0.996**; 736 L, 0.855; 737 I, 0.803; 751 N, 0.760; 809 A, 0.828; 906 N, 0.768; 914 E, 0.795; 915 L, 0.788; 929 L, 0.835; 957 K, 0.979*; 1030 E, 0.979*; 1116 K, 0.804; 1172 M, 0.731; 1186 Q, 0.798; 1206 Q, 0.798; 1502 Q, 0.754; 1601 I, 1.000**; 1616 D, 0.773;
1631 R, 0.814; 1652 R, 0.795; 1665 V, 0.738; 1685 E, 0.978*; 1710 I, 0.979*; 1739 R, 0.793ycf2 14.95 5.7×10-4 14.95 5.7×10-4 30 R, 0.767; 56 R, 0.770; 103 L, 0.765; 126 S, 0.765; 248 S, 0.769; 285 L, 0.765; 320 L, 0.766; 332 R, 0.764; 432 V, 0.765; 466 Y, 0.765; 573 I, 0.764; 598 L, 0.766; 606 M, 0.759; 759 F, 0.763; 850 S, 0.965*; 943 E, 0.764; 1037 P, 0.766; 1050 F, 0.765; 1064 S, 0.763; 1074 I, 0.965*; 1093 L, 0.767; 1101 N, 0.964*; 1171 W, 0.767; 1173 S, 0.765; 1245 P, 0.766; 1265 D, 0.762; 1309 D, 0.762; 1344 R, 0.764; 1422 L, 0.965*; 1431 F, 0.763; 1522 D, 0.767; 1550 Y, 0.766; 1656 N, 0.763; 1717 S, 0.768; 1799 R, 0.769; 2045 E, 0.764 *和**分别表示正选择位点概率> 95%和> 99%。
* and ** indicate that the probabilities of positively selected sites are > 95% and > 99%, respectively.2.6 人参属系统发育分析
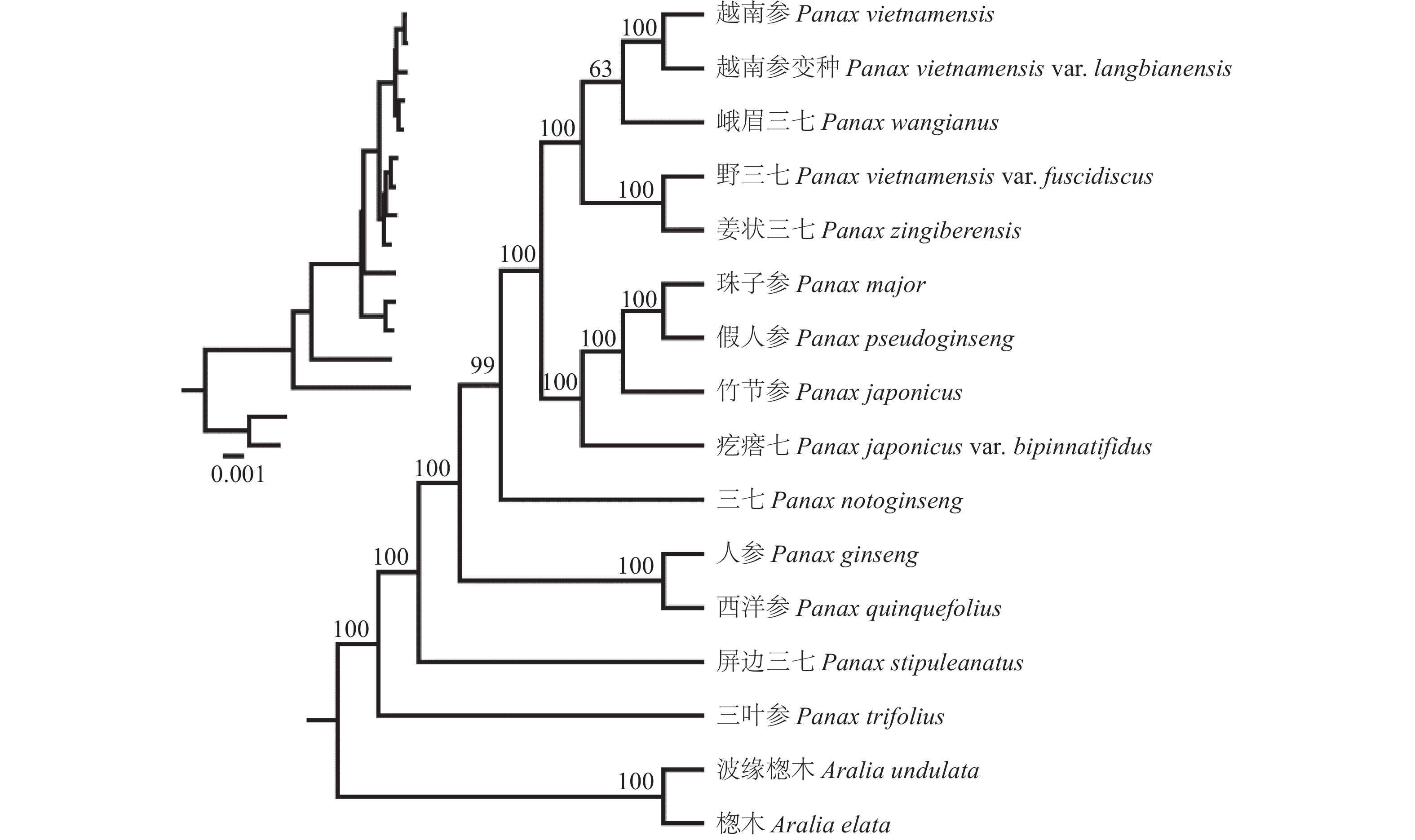
基于叶绿素基因组数据,使用IQ-TREE软件构建了人参属物种系统发育树(图7)。结果显示,屏边三七和三叶参位于人参属系统发育树的基部,人参和西洋参构成支持率为100%的单一支系。三七单独构成一支。越南参及其两个变种与峨眉三七和姜状三七构成一支,与由珠子参、假人参、竹节参和疙瘩七构成的单支形成姊妹类群。
3. 讨论
人参属植物是一类生长缓慢的多年生草本植物,根茎中富含人参皂苷等生物活性物质,被广泛用于预防和治疗多种疾病[5]。目前,多个人参属物种叶绿体基因组已完成测序[2,13-14]。通过叶绿体基因组分析,可深入探讨人参属物种的系统发育与进化特征,为该属种质资源保护、分子育种和品种改良奠定基础。本研究从NCBI数据库下载人参属物种叶绿体基因组数据,对基因组基因构成、序列重复、结构变异、基因进化及物种系统发育关系进行分析,发现14种人参属植物叶绿体基因组高度保守,物种间叶绿体基因组大小差异500 bp以内,编码的基因种类、数目和排序一致,显示基因组结构高度保守。
物种间的重复序列差异是进化过程中自然选择和环境适应性的结果,重复序列数量与物种的进化水平直接相关[23]。长重复序列广泛存在于植物叶绿体基因组中,其通过保护编码区维持了基因组的稳定性,在基因表达和调控中发挥重要作用,由于具有高度多态性,长重复序列成为群体遗传学研究的理想工具[24]。人参属物种叶绿体基因组中平均包含24条回文重复和20条正向重复,而反向重复和互补重复较少,30~39 bp长度的重复序列占50%以上,这与其他物种类似[25]。人参属重复序列数目高于木姜子属(Litsea)[8],低于辣椒属(Capsicum)[10]、悬钩子属(Rubus)[11]、姜属(Zingiber)[26]和冬青属(Ilex)[27]。与人参属其他物种(除越南参变种基因组较小外)相比,屏边三七和三叶参均含有较少的重复序列,这在一定程度上反映物种进化水平,与二者属于基部类群的结果一致。简单序列重复具有丰富的多态性、高度重复性和可靠性等优点,特别是在种内水平上表现出相对较高的突变率,在群体遗传关系和系统发育研究中经常被用作遗传分子标记[28],被广泛用于药用植物群体遗传学研究[10-11]。人参属物种叶绿体基因组中含有37~44个SSR位点,其中大多数为A/T重复,单核苷酸重复是最丰富的重复序列,与其他物种研究类似[8,10,29-30],这与植物叶绿体基因组A、T含量较高有关。人参属物种SSR数目高于悬钩子属[11]和松属(Pinus)[24],低于姜属[26]和草果(Amomum tsao-ko)[31]。本研究中发现的长重复序列以及SSR将有助于开发人参属物种的遗传多态性分子标记,为群体遗传学研究提供材料。
IR区边界的收缩和扩张是叶绿体基因组结构变异的重要原因[32]。人参属叶绿体基因组IR区与单拷贝区边界高度保守[13],4个边界分别跨越rps19、ycf1-ndhF、ycf1和trnH,但不同物种 IR/SSC边界的差异也较为显著,IR/SSC边界的变化可能是导致人参属叶绿体间大小差异的原因,这与五加科其他物种类似[33]。叶绿体基因组共线性分析显示,物种演化过程中人参属叶绿体基因组未发生基因重排,IR区序列一致性高于单拷贝区,这与前人研究结果一致[3,11,34]。通过比较14种人参属植物叶绿体基因组,检测到12个高度可变热点,7个位于LSC区(trnH-psbA、rps16-trnQ、psbI-trnS、trnE-trnT、clpP、psbH-petB和rpl16-rps3),5个位于SSC区(ndhF-rpl32、rpl32-trnL、ndhD-psaC、rps4-trnT和ycf1),IR区未检测到高变位点,表明单拷贝区核苷酸多态性明显高于IR区,且多态性位点多为基因间隔区[35-36]。鉴定的12个高变区可与SSR等其他标记一起用于群体遗传学和系统地理学研究[11,30]。
植物在进化过程中,往往受到环境等多种因素的影响,而基因的正选择分析可为植物适应性进化提供理论依据。当dN/dS>1,认为基因主要受正选择作用;dN/dS=1,认为主要受中性选择作用;dN/dS<1,则认为主要受纯化选择作用[37]。研究发现,人参属叶绿体功能未知基因clpP、ycf1和ycf2受正选择作用。目前,clpP、ycf1和ycf2编码蛋白功能仍不明确,但其基因在多个物种中均受正选择作用[25-26,31]。与本研究结果不同,Liu等[13]对8个人参属物种分析后,未检测到正选择位点,这可能与物种数目、检测软件和计算方法有关。本研究使用14种人参属植物的基因序列,采取通用的CODEML算法,能得到更准确的结果。在光合作用相关基因ndhF和rbcL中检测到多个正选择位点,可能与人参属物种不同纬度生长环境的光照条件有关。以上分析表明,这些正选择基因或位点在人参属植物与环境互作过程中受到了选择作用,可能与物种的生态适应性有关。
基于完整叶绿体基因组序列的人参属系统发育分析显示,屏边三七和三叶参位于基部分支,这与前人研究结果一致[2,14,38]。与本研究不同,岳杰[14]认为珠子参与越南参和姜状三七关系更为紧密,这可能与建树时选择的基因组序列不同有关,NCBI数据库中有3条珠子参叶绿体基因组序列,本研究选择了聚类关系较近的两条序列中的一条(MN496312)。本研究结果中的物种系统演化拓扑结构与前人研究类似,人参和西洋参在进化过程中经历了两次全基因组复制事件,促进了人参属物种的多样化[6,39]。人参和西洋参四倍化后于2.59百万年前从二倍体物种中分化出来,主要分布在东北亚和北美,由于地理隔离和生态环境适应性进化,与其他二倍体物种进一步分开,于0.77百万年前分离成单种[6]。
4. 结论
人参属植物叶绿体基因组长度为155 984 ~156 402 bp,均包含114个unique基因,其中80个蛋白编码基因,4个rRNA和30个tRNA。基因组序列中含有较多的回文重复和正向重复。SSR主要由A/T碱基构成,以单核苷酸重复为主。人参属植物叶绿体基因组序列一致性较高,非编码区序列变异高于编码区,IR区存在一定程度的扩张和收缩。12个高度可变热点中,7个位于LSC区,5个位于SSC区。未知功能基因clpP、ycf1和ycf2受正选择作用。屏边三七和三叶参为人参属的原始类群,而三七、竹节参和越南参则亲缘关系较近,构成了人参属的较新分化类群。
-
表 1 人参属物种叶绿体基因组特征
Table 1 Chloroplast genomes of Panax species
物种 Species 登录号 Accession 长度 Length/bp GC含量 GC content/% 基因组 Genome LSC SSC IR 基因组 Genome LSC SSC IR 人参
P. ginsengMK408938 156 241 86 128 18 077 26 018 38.07 36.27 32.18 43.10 竹节参
P. japonicusKP036469 156 188 86 199 18 013 25 988 38.07 36.29 32.21 43.05 疙瘩七
P. japonicus var. bipinnatifidusMK408962 156 244 86 186 18 006 26 026 38.06 36.28 32.21 43.05 珠子参
P. majorMN496312 156 402 86 189 18 007 26 103 38.07 36.28 32.22 43.05 三七
P. notoginsengMK408945 156 319 86 157 18 004 26 079 38.07 36.27 32.23 43.07 假人参
P. pseudoginsengMW145449 156 074 86 124 18 008 25 971 38.06 36.27 32.22 43.06 西洋参
P. quinquefoliusMK408953 156 070 86 077 17 993 26 000 38.07 36.27 32.22 43.08 屏边三七
P. stipuleanatusMK408965 156 007 86 083 18 150 25 887 38.04 36.26 32.07 43.08 三叶参
P. trifoliusMF100782 156 157 86 322 18 047 25 894 38.08 36.27 32.26 43.10 越南参
P. vietnamensisKP036470 155 993 86 178 17 935 25 940 38.05 36.27 32.22 43.02 野三七
P. vietnamensis var. fuscidiscusMT798585 156 284 86 171 17 971 26 071 38.06 36.28 32.27 42.98 越南参变种
P. vietnamensis var. langbianensisMT798583 155 984 86 174 17 934 25 938 38.05 36.27 32.21 43.03 峨眉三七
P. wangianusMK408964 156 189 86 190 17 969 26 015 38.07 36.29 32.21 43.03 姜状三七
P. zingiberensisMK408969 156 192 86 116 17 966 26 055 38.07 36.31 32.27 43.00 表 2 基于位点模型的人参属叶绿体蛋白编码基因正选择分析
Table 2 Potential positive selection test on chloroplast genomes of Panax species based on site model
基因
GeneM1 vs M2 M7 vs M8 正选择位点
Positively selected sites2ΔlnL P 值 2ΔlnL P 值 accD 2.39 3.0×10-1 2.47 2.9×10-1 139 Q, 0.505; 141 S, 0.517; 144 R, 0.528; 209 A, 0.856; 264 E, 0.504; 364 I, 0.507; 372 L, 0.526; 494 Q, 0.855 atpF 3.37 1.9×10-1 3.42 1.8×10-1 14 W, 0.541; 77 Q, 0.537; 158 G, 0.854 cemA 7.48 2.4×10-2 7.48 2.4×10-2 14 A, 0.547; 22 W, 0.538; 54 K, 0.536; 102 D, 0.533; 103 R, 0.544; 118 V, 0.973*; 205 F, 0.531; 208 W, 0.540 clpP 13.40 1.2×10-3 13.40 1.2×10-3 38 P, 0.990*; 39 V, 0.622; 40 A, 0.935; 41 S, 0.612; 138 G, 0.629; 160 Q, 0.626 matK 19.22 6.7×10-5 19.25 6.6×10-5 83 R, 0.994** ndhD 0.77 6.8×10-1 0.99 6.1×10-1 22 F, 0.830; 41 I, 0.829 ndhF 19.81 5.0×10-5 20.29 3.9×10-5 17 P, 0.597; 123 R, 0.592; 133 T, 0.583; 223 F, 0.959*; 421 I, 0.964*; 463 Q, 0.591; 483 R, 0.569; 499 H, 0.552; 502 A, 0.965*; 511 Q, 0.591; 512 M, 0.526; 532 K, 0.559; 534 I, 0.962*; 572 N, 0.543; 598 D, 0.573; 606 F, 0.548; 621 L, 0.630; 641 I, 0.549; 642 G, 0.597; 657 G, 0.594; 670 A, 0.963*; 677 I, 0.532; 703 S, 0.548; 737 Y, 0.964*; 743 F, 0.537; 745 D, 0.958*; 746 L, 1.000** rbcL 15.94 3.5×10-4 16.10 3.2×10-4 225 L, 0.632; 226 Y, 0.969*; 230 A, 0.635; 270 I, 0.866; 281 A, 0.644; 328 S, 0.999**; 353 F, 0.584; 439 R, 0.968*; 472 V, 0.609; 474 I, 0.621; 478 V, 0.714 rpl2 0 1 0 1 99 K, 0.651; 265 D, 0.650; 267 L, 0.652 rpoA 14.53 7.0×10-4 14.56 6.9×10-4 34 L, 0.506; 256 E, 0.991**; 264 N, 0.999**; 272 S, 0.533; 320 A, 0.849; 337 L, 0.537 rpoB 0.90 6.4×10-1 0.90 6.4×10-1 37 Y, 0.505; 165 R, 0.502; 193 R, 0.502; 295 I, 0.502; 365 T, 0.504; 459 S, 0.502; 695 K, 0.506; 735 T, 0.504; 935 D, 0.832 rpoC2 0 1 0 1 85 Q, 0.736 rps12 0 1 0.02 9.9×10-1 25 R, 0.502 ycf1 75.81 0 75.78 0 205 K, 1.000**; 206 Y, 0.999**; 288 I, 0.876; 532 T, 0.803; 533 K, 0.956*; 604 R, 0.801; 624 R, 0.825; 668 I, 0.779; 685 W, 0.837; 699 V, 0.824; 701 Q, 0.802; 706 V, 0.814; 720 V, 0.856; 722 T, 0.814; 723 D, 0.808; 731 R, 0.982*; 733 K, 0.809; 734 I, 0.996**; 736 L, 0.855; 737 I, 0.803; 751 N, 0.760; 809 A, 0.828; 906 N, 0.768; 914 E, 0.795; 915 L, 0.788; 929 L, 0.835; 957 K, 0.979*; 1030 E, 0.979*; 1116 K, 0.804; 1172 M, 0.731; 1186 Q, 0.798; 1206 Q, 0.798; 1502 Q, 0.754; 1601 I, 1.000**; 1616 D, 0.773;
1631 R, 0.814; 1652 R, 0.795; 1665 V, 0.738; 1685 E, 0.978*; 1710 I, 0.979*; 1739 R, 0.793ycf2 14.95 5.7×10-4 14.95 5.7×10-4 30 R, 0.767; 56 R, 0.770; 103 L, 0.765; 126 S, 0.765; 248 S, 0.769; 285 L, 0.765; 320 L, 0.766; 332 R, 0.764; 432 V, 0.765; 466 Y, 0.765; 573 I, 0.764; 598 L, 0.766; 606 M, 0.759; 759 F, 0.763; 850 S, 0.965*; 943 E, 0.764; 1037 P, 0.766; 1050 F, 0.765; 1064 S, 0.763; 1074 I, 0.965*; 1093 L, 0.767; 1101 N, 0.964*; 1171 W, 0.767; 1173 S, 0.765; 1245 P, 0.766; 1265 D, 0.762; 1309 D, 0.762; 1344 R, 0.764; 1422 L, 0.965*; 1431 F, 0.763; 1522 D, 0.767; 1550 Y, 0.766; 1656 N, 0.763; 1717 S, 0.768; 1799 R, 0.769; 2045 E, 0.764 *和**分别表示正选择位点概率> 95%和> 99%。
* and ** indicate that the probabilities of positively selected sites are > 95% and > 99%, respectively. -
[1] WEN J, ZIMMER E A. Phylogeny and biogeography of Panax L. (The Ginseng Genus, Araliaceae): Inferences from ITS sequences of nuclear ribosomal DNA [J]. Molecular Phylogenetics and Evolution, 1996, 6(2): 167−177. DOI: 10.1006/mpev.1996.0069
[2] NGUYEN V B, LINH GIANG V N, WAMINAL N E, et al. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers [J]. Journal of Ginseng Research, 2020, 41(1): 135−144. DOI: 10.1016/j.jgr.2018.06.003
[3] NGUYEN V B, PARK H S, LEE S C, et al. Authentication markers for five major Panax species developed via comparative analysis of complete chloroplast genome sequences [J]. Journal of Agricultural and Food Chemistry, 2017, 65(30): 6298−6306. DOI: 10.1021/acs.jafc.7b00925
[4] LI S, WANG P, YANG W Z, et al. Characterization of the components and pharmacological effects of mountain-cultivated ginseng and garden ginseng based on the integrative pharmacology strategy [J]. Frontiers in Pharmacology, 2021, 12: 659954. DOI: 10.3389/fphar.2021.659954
[5] SHIN B K, KWON S W, PARK J H. Chemical diversity of ginseng saponins from Panax ginseng [J]. Journal of Ginseng Research, 2015, 39(4): 287−298. DOI: 10.1016/j.jgr.2014.12.005
[6] CHOI H I, KIM N H, LEE J, et al. Evolutionary relationship of Panax ginseng and P. quinquefolius inferred from sequencing and comparative analysis of expressed sequence tags [J]. Genetic Resources and Crop Evolution, 2013, 60(4): 1377−1387. DOI: 10.1007/s10722-012-9926-3
[7] YANG Z J, LIU G Z, ZHANG G H, et al. The chromosome-scale high-quality genome assembly of Panax notoginseng provides insight into dencichine biosynthesis [J]. Plant Biotechnology Journal, 2021, 19(5): 869−871. DOI: 10.1111/pbi.13558
[8] LIU C, CHEN H H, TANG L Z, et al. Plastid genome evolution of a monophyletic group in the subtribe Lauriineae (Laureae, Lauraceae)[J]. Plant Diversity, 2022 , 44 (4): 377 − 388. DOI: 10.1016/j.pld.2021.11.009
[9] 刘潮, 韩利红, 彭悦, 等. 黄丹木姜子叶绿体基因组特征分析 [J]. 南方农业学报, 2022, 53(1):12−20. DOI: 10.3969/j.issn.2095-1191.2022.01.002 LIU C, HAN L H, PENG Y, et al. Characteristics of chloroplast genome of Litsea elongata(wall. ex nees)Benth. et hook. F [J]. Journal of Southern Agriculture, 2022, 53(1): 12−20.(in Chinese) DOI: 10.3969/j.issn.2095-1191.2022.01.002
[10] 刘潮, 韩利红, 代小波, 等. 辣椒属叶绿体基因组特征及进化 [J]. 热带作物学报, 2022, 43(3):447−454. DOI: 10.3969/j.issn.1000-2561.2022.03.002 LIU C, HAN L H, DAI X B, et al. Characteristics and phylogenetics of the complete chloroplast genomes of Capsicum species [J]. Chinese Journal of Tropical Crops, 2022, 43(3): 447−454.(in Chinese) DOI: 10.3969/j.issn.1000-2561.2022.03.002
[11] YU J J, FU J, FANG Y P, et al. Complete chloroplast genomes of Rubus species (Rosaceae) and comparative analysis within the genus [J]. BMC Genomics, 2022, 23(1): 32. DOI: 10.1186/s12864-021-08225-6
[12] 刘潮, 唐利洲, 韩利红. 四川山胡椒叶绿体基因组特征及山胡椒属系统发育 [J]. 林业科学, 2021, 57(12):167−174. LIU C, TANG L Z, HAN L H. Characterization of the chloroplast genome of Lindera setchuenensis and phylogenetics of the genus Lindera [J]. Scientia Silvae Sinicae, 2021, 57(12): 167−174.(in Chinese)
[13] LIU C K, YANG Z Y, YANG L F, et al. The complete plastome of Panax stipuleanatus: Comparative and phylogenetic analyses of the genus Panax (Araliaceae) [J]. Plant Diversity, 2018, 40(6): 265−276. DOI: 10.1016/j.pld.2018.11.001
[14] 岳杰. 四种人参属植物叶绿体全基因组结构及其系统发育分析[D]. 汉中: 陕西理工大学, 2021 YUE J. Structural and phylogenetic analyses of the complete chloroplast genomes of four species within the genus Panax linn. [D]. Hanzhong: Shaanxi University of Technology, 2021. (in Chinese)
[15] KURTZ S, CHOUDHURI J V, OHLEBUSCH E, et al. REPuter: The manifold applications of repeat analysis on a genomic scale [J]. Nucleic Acids Research, 2001, 29(22): 4633−4642. DOI: 10.1093/nar/29.22.4633
[16] BEIER S, THIEL T, MÜNCH T, et al. MISA-web: A web server for microsatellite prediction [J]. Bioinformatics, 2017, 33(16): 2583−2585. DOI: 10.1093/bioinformatics/btx198
[17] AMIRYOUSEFI A, HYVÖNEN J, POCZAI P. IRscope: An online program to visualize the junction sites of chloroplast genomes [J]. Bioinformatics (Oxford, England), 2018, 34(17): 3030−3031. DOI: 10.1093/bioinformatics/bty220
[18] FRAZER K A, PACHTER L, POLIAKOV A, et al. VISTA: Computational tools for comparative genomics[J]. Nucleic Acids Research, 2004, 32(Web Server issue): W273-W279.
[19] KATOH K, ROZEWICKI J, YAMADA K D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization [J]. Briefings in Bioinformatics, 2019, 20(4): 1160−1166. DOI: 10.1093/bib/bbx108
[20] ROZAS J, FERRER-MATA A, SÁNCHEZ-DELBARRIO J C, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets [J]. Molecular Biology and Evolution, 2017, 34(12): 3299−3302. DOI: 10.1093/molbev/msx248
[21] XU B, YANG Z H. PAMLX: A graphical user interface for PAML [J]. Molecular Biology and Evolution, 2013, 30(12): 2723−2724. DOI: 10.1093/molbev/mst179
[22] MINH B Q, SCHMIDT H A, CHERNOMOR O, et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era [J]. Molecular Biology and Evolution, 2020, 37(5): 1530−1534. DOI: 10.1093/molbev/msaa015
[23] GANLEY A R D, KOBAYASHI T. Monitoring the rate and dynamics of concerted evolution in the ribosomal DNA repeats of Saccharomyces cerevisiae using experimental evolution [J]. Molecular Biology and Evolution, 2011, 28(10): 2883−2891. DOI: 10.1093/molbev/msr117
[24] ZEB U, DONG W L, ZHANG T T, et al. Comparative plastid genomics of Pinus species: Insights into sequence variations and phylogenetic relationships [J]. Journal of Systematics and Evolution, 2020, 58(2): 118−132. DOI: 10.1111/jse.12492
[25] ZHU B, QIAN F, HOU Y F, et al. Complete chloroplast genome features and phylogenetic analysis of Eruca sativa (Brassicaceae) [J]. PLoS One, 2021, 16(3): e0248556. DOI: 10.1371/journal.pone.0248556
[26] LI D M, YE Y J, XU Y C, et al. Complete chloroplast genomes of Zingiber montanum and Zingiber zerumbet: Genome structure, comparative and phylogenetic analyses [J]. PLoS One, 2020, 15(7): e0236590. DOI: 10.1371/journal.pone.0236590
[27] XU K W, LIN C X, LEE S Y, et al. Comparative analysis of complete Ilex (Aquifoliaceae) chloroplast genomes: Insights into evolutionary dynamics and phylogenetic relationships [J]. BMC Genomics, 2022, 23(1): 203. DOI: 10.1186/s12864-022-08397-9
[28] KASHI Y, KING D G. Simple sequence repeats as advantageous mutators in evolution [J]. Trends in Genetics, 2006, 22(5): 253−259. DOI: 10.1016/j.tig.2006.03.005
[29] HAN C Y, DING R, ZONG X Y, et al. Structural characterization of Platanthera ussuriensis chloroplast genome and comparative analyses with other species of Orchidaceae [J]. BMC Genomics, 2022, 23(1): 84. DOI: 10.1186/s12864-022-08319-9
[30] YAN J W, LI J H, YU L, et al. Comparative chloroplast genomes of Prunus subgenus Cerasus (Rosaceae): Insights into sequence variations and phylogenetic relationships [J]. Tree Genetics & Genomes, 2021, 17(6): 50.
[31] 马孟莉, 张薇, 孟衡玲, 等. 草果叶绿体基因组特征及系统发育分析 [J]. 中草药, 2021, 52(19):6023−6031. DOI: 10.7501/j.issn.0253-2670.2021.19.025 MA M L, ZHANG W, MENG H L, et al. Characterization and phylogenetic analysis of the complete chloroplast genome of Amomum tsao-ko [J]. Chinese Traditional and Herbal Drugs, 2021, 52(19): 6023−6031.(in Chinese) DOI: 10.7501/j.issn.0253-2670.2021.19.025
[32] KIM K J. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng nees) and comparative analysis of sequence evolution among 17 vascular plants [J]. DNA Research, 2004, 11(4): 247−261. DOI: 10.1093/dnares/11.4.247
[33] LI R, MA P F, WEN J, et al. Complete sequencing of five Araliaceae chloroplast genomes and the phylogenetic implications [J]. PLoS One, 2013, 8(10): e78568. DOI: 10.1371/journal.pone.0078568
[34] TROFIMOV D, CADAR D, SCHMIDT-CHANASIT J, et al. A comparative analysis of complete chloroplast genomes of seven Ocotea species (Lauraceae) confirms low sequence divergence within the Ocotea complex [J]. Scientific Reports, 2022, 12: 1120. DOI: 10.1038/s41598-021-04635-4
[35] YANG J, CHIANG Y C, HSU T W, et al. Characterization and comparative analysis among plastome sequences of eight endemic Rubus (Rosaceae) species in Taiwan [J]. Scientific Reports, 2021, 11: 1152. DOI: 10.1038/s41598-020-80143-1
[36] NIU Y F, GAO C W, LIU J. Comparative analysis of the complete plastid genomes of Mangifera species and gene transfer between plastid and mitochondrial genomes [J]. PeerJ, 2021, 9: e10774. DOI: 10.7717/peerj.10774
[37] GARUD N R, POLLARD K S. Population genetics in the human microbiome [J]. Trends in Genetics:TIG, 2020, 36(1): 53−67. DOI: 10.1016/j.tig.2019.10.010
[38] JI Y H, LIU C K, YANG Z Y, et al. Testing and using complete plastomes and ribosomal DNA sequences as the next generation DNA barcodes in Panax (Araliaceae) [J]. Molecular Ecology Resources, 2019, 19(5): 1333−1345. DOI: 10.1111/1755-0998.13050
[39] KIM N H, CHOI H I, KIM K H, et al. Evidence of genome duplication revealed by sequence analysis of multi-loci expressed sequence tag-simple sequence repeat bands in Panax ginseng Meyer [J]. Journal of Ginseng Research, 2014, 38(2): 130−135. DOI: 10.1016/j.jgr.2013.12.005
-
期刊类型引用(3)
1. 李启少,王正德,李建运,王飞,李子涵,朱文,汤嘉莹,辛雅萱,李斌,辛培尧. 棕红悬钩子叶绿体的基因组结构与序列特征分析. 西南林业大学学报(自然科学). 2024(04): 93-105 .  百度学术
百度学术
2. 彭毅丹,马楠,叶奕含,刘亦欣,谭启怡,陈莹,张雨曲,杨新杰. 秦巴山区三种人参属药用植物叶绿体基因组特征分析. 广西植物. 2024(06): 1138-1150 .  百度学术
百度学术
3. 李春连,张鹏,蔡楚平,覃水凤,肖善芳,罗秋香,郭松,蓝峻峰. 瑶药鸭脚风叶绿体基因组结构解析及密码子偏好性分析. 中国农业科技导报. 2024(12): 63-76 .  百度学术
百度学术
其他类型引用(0)




 下载:
下载: