Genetic Diversity and Principal Components of Tea Germplasms from Shiting Plantations in Nan'an
-
摘要:目的
深入了解南安石亭群体种茶树种质资源(Camellia sinensis)的遗传多样性及其直接利用价值,为南安石亭群体种茶树种质资源的创新利用提供科学依据。
方法利用SNP分子标记技术对17份南安石亭群体种茶树种质资源及福建6个代表性品种进行亲缘关系及遗传多样性分析。进一步采用高效液相色谱法(HPLC)和超高效液相色谱-串联质谱法(UPLC-MS/MS)测定代表性的10份南安石亭群体种茶树种质资源儿茶素及游离氨基酸等组分含量。
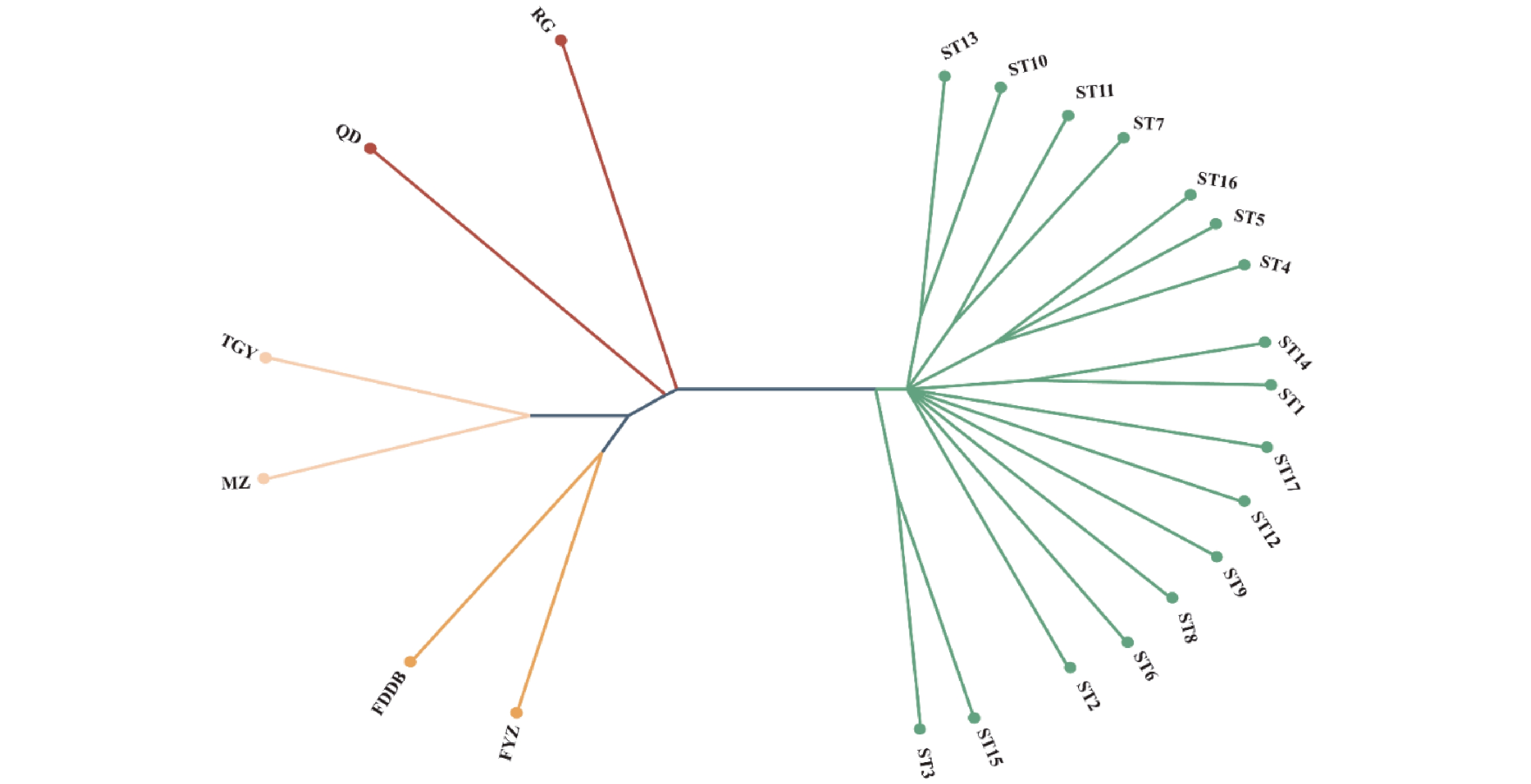
结果筛选出44个适用于鉴定南安石亭群体种茶树种质资源的SNP位点,构建南安石亭群体种茶树种质资源SNP指纹图谱。通过UPGMA进化树构建,发现23个样品可分为4个亚群,南安石亭群体种茶树种质资源与闽北地区的肉桂、奇丹亲缘关系较近。此外,进行了氨基酸,儿茶素含量分析,发现南安石亭群体种茶树种质资源的主要品质成分含量差异显著(P<0.05)。儿茶素总量为107.55~177.47 mg·g−1,游离氨基酸总量为8.35~32.32 mg·g−1。其中ST2、ST3、ST6、ST17种质具有较高的EGCG或EGCG3"Me含量,ST3种质氨基酸含量最高、儿茶素苦涩味指数最低。
结论南安石亭群体种茶树种质资源丰富,与闽北地区茶树资源亲缘关系较为亲密。ST2、ST3、ST6和ST17具有较高的EGCG或EGCG3"Me含量,ST3适制绿茶,可进一步开展选育工作。
Abstract:ObjectiveGenetic diversity of the tea germplasms preserved at Shiting plantations in Nan’an was studied for promoting the utilization of the local specialty.
MethodsSNP molecular marker technology was applied to analyze the genetic diversity and relationships of 17 tea germplasms from the Shiting plantations and 6 representative varieties from Fujian. HPLC and UPLC-MS/MS were employed to determine the contents of catechins and free amino acids in 10 selected samples for a comparison among the tea varieties.
ResultsForty-four SNP loci suitable for identifying the tea plants were selected. An SNP fingerprint profile of the germplasms was constructed. The UPGMA evolutionary tree divided the 23 germplasms into 4 subgroups, which showed a close relation with Rougui and Qidan teas from northern Fujian. Significant differences were found on the contents of major amino acids and catechins among the varieties (P<0.05) with total free amino acids from 8.46 mg·g−1 to 32.66 mg·g−1 and catechins ranging from 107.56 mg·g−1 to 177.60 mg·g−1. The germplasms coded ST2, ST3, ST6, and ST17 had high contents of EGCG or EGCG3"Me, while ST3 the most on amino acids and the least on catechin bitterness index.
ConclusionShiting plantations in Nan’an had a rich collection of tea germplasms that were closely related to those in northern Fujian. Among them, ST2, ST3, ST6, and ST17 teas had high contents of either EGCG or EGCG3"Me. ST3, specifically, appeared to be suitable for making green tea as well as in breeding programs.
-
Keywords:
- Shiting, Nan’an /
- Camellia sinensis /
- germplasm /
- genetic diversity /
- component analysis /
- SNP
-
0. 引言
【研究意义】粘质沙雷氏菌(Serratia marcescens)是肠杆菌科中的一类革兰氏阴性短杆状细菌,该菌在土壤、水体等环境中广泛分布[1]。粘质沙雷氏菌发现于1819年,20世纪70年代确认该种中某些菌株为条件致病菌[2]。近年陆续发现,粘质沙雷氏菌可以产生多种有潜力的次生代谢物质,如肽类抗生素,生物表面活性剂serrawettin W1和灵菌红素[3-5]等。该菌也可作为底盘微生物高产N-乙酰神经氨酸[6]、乙偶姻和丁二醇等高附加值产品[7-8]。因此,粘质沙雷氏菌已成为有重要前景的工业微生物菌种,研究其产物合成及调控相关基因以揭示其产物合成机制有助于挖掘其生产潜力。【前人研究进展】灵菌红素类物质由于具有抗菌和抗肿瘤等生物活性而成为近年研究热点,粘质沙雷氏菌是特异性产灵菌红素的最主要菌种[9]。研究表明,粘质沙雷氏菌合成灵菌红素受多个基因调控,包括细菌群体感应相关基因,双组分系统相关基因和其他调控基因[10]。双组分调控系统广泛存在于细菌中,参与压力响应、细菌形态、 运动性、 菌毛合成及毒力表现等[11],典型的双组分系统由作为传感器的跨膜组氨酸激酶和作为效应分子的一个细胞质蛋白组成[12]。目前已知双组分系统PigQ/PigW、PhoB/PhoR和EepR/EepS正调控粘质沙雷氏菌灵菌红素的合成[13-15],双组分系统RssB/RssA [16]对灵菌红素的合成起到负调控作用。更多灵菌红素合成相关调控基因的揭示有助于理解该类菌株合成灵菌红素的调控机制和规律。【本研究切入点】EnvZ/OmpR是一个典型的双组分调控系统,但是其对粘质沙雷氏菌灵菌红素合成及其他生物学特性的影响知之甚少。【拟解决的关键问题】利用从一株高产灵菌红素粘质沙雷氏菌FZSF02菌株[17]的Tn5转座子突变体库中获得一株ompR突变株,检测ompR敲除对菌株生物学特性的影响,初步揭示EnvZ/OmpR双组分调控系统对粘质沙雷氏菌的作用。
1. 材料与方法
1.1 试验材料
1.1.1 菌株和质粒
大肠杆菌DH5α由本实验室保存,粘质沙雷氏菌FZSF02由本实验室自土壤中分离,粘质沙雷氏菌FZSF02ompR:: Tn5源自本实验室由FZSF02构建的Tn5转座子突变体库。
1.1.2 主要试剂和仪器
供试培养基及抗生素购自生工生物工程(上海)股份有限公司,高保真DNA聚合酶PrimeSTAR® Max DNA Polymerase(Takara,R045A),克隆试剂盒Zero Background pTOPO-Blunt Simple Cloning Kit购自北京艾德莱生物科技有限公司,转座子突变试剂盒EZ-Tn5™ <KAN-2>Tnp Transposome™ Kit (TSM99K2, Epicentre),细菌总RNA提取试剂盒(TIANGEN,DP430)购自天根生化科技(北京)有限公司,琼脂粉(BD:Becton, Dickinson and Company),吐温80(国药集团),脱脂奶粉(BD:Becton, Dickinson and Company),血琼脂平板(青岛海博生物),96孔细胞培养板(Greiner Bio-One,No.655180),美国MD SpectraMax190全波长酶标仪,电转化仪Gene Pulser Xcell(Bio-Rad)。
1.2 试验方法
1.2.1 目的基因的序列分析
调取目的基因的编码蛋白序列,通过NCBI(National Center for Biotechnology Information)网站中blastp功能比对查找与目的蛋白同源的蛋白序列并预测蛋白的功能,挑选隶属不同种属的同源蛋白序列,应用MEGA5.0软件Neighbor-joining法构建该蛋白的系统发育树。
1.2.2 电击转化感受态细胞的制备
LB琼脂平板上挑取FZSF02菌株1个单菌落置于50 mL液体LB培养基中,37 ℃、180 r·min−1培养12 h作为种子液,以1% (v/v)的接种量转接至50 mL液体LB培养基中37 ℃,180 r·min−1培养至OD600约1.0时将盛有菌液的摇瓶冰浴30 min,冰浴结束后转移至50 mL离心管于4 ℃离心机中5 000 r·min−1离心10 min;弃上清后用40 mL预冷的无菌水洗涤后4 ℃离心10 min,重复用无菌水洗涤离心1次;用40 mL的冰预冷的10%甘油洗涤后4%离心10 min,重复用10%甘油洗涤离心一次;用0.5 mL,10%甘油悬浮菌体沉淀,以 100 μL每管分装至1.5 mL离心管后保存在−80 ℃备用。
1.2.3 突变株及敲除菌株的获得
ompR突变株的构建应用转座子突变试剂盒EZ-Tn5™ <KAN-2>Tnp Transposome™ Kit (TSM99K2, Epicentre)。转座子插入位置的鉴定应用High TAIL-PCR [18-19]。ompR基因的敲除应用同源重组法,扩增含有启动子的卡那抗性基因,应用ompR前端引物OmpRFF和OmpRFR扩增ompR上游同源臂,应用OmpRBF和OmpRBR扩增ompR基因的下游同源臂,将3段序列通过重叠PCR(Overlap PCR)拼接后连接Zero Background pTOPO-Blunt质粒,在氨苄青霉素和卡那霉素双抗性LB琼脂平板上筛选阳性克隆子。以阳性克隆子质粒为模板应用引物OmpRFF和OmpRBR扩增获得含有ompR上游同源臂、卡那霉素抗性基因和ompR下游同源臂的DNA片段,片段经纯化后用灭菌超纯水洗脱。上述PCR扩增所用引物详细序列见表1。通过电击转化将上述DNA片段转化FZSF02野生型菌株,电转参数为:25 μF, 200 ohm, 1 800 V。在卡那霉素质量浓度为100 mg·L−1的LB琼脂平板上挑取单克隆验证。
表 1 供试引物Table 1. Primers used in this study引物 Primer 序列 Sequence(5′-3′) OmpRFF ATGCAAGAGAATCATAAGATCCTG OmpRFR CATCGATGATGGTTGAGAGTCGGCGCCGATTTCCAGCCCCA OmpRBF CTCGATGAGTTTTTCTAAGGCAAATTCAAACTGAACCTCGGC OmpRBR TCATGCCTTGCTGCCGTCCGGTAC KanF TCTCAACCATCATCGATGAATTGT KanR TTAGAAAAACTCATCGAGCATCAA pigAF CGCCATCTTCCACGATTCAA pigAR CATTAGCCGACACTGTTCCA pigFF CACGGTATTCGGCGATGAC pigFR CACGGTGTTGCGAGAAGT pigNF CGGTTACCCTGGTCTATTG pigNR TGTCAGCACGATGTTCAT 16SF CGTTACTCGCAGAAGAAGCA 16SR TCACCGCTACACCTGGAA 1.2.4 细菌生长曲线测定
挑单菌落至50 mL液体培养基中,37 ℃、180 r·min−1培养FZSF02野生型菌株WT和基因敲除菌株ΔompR过夜作为种子液,调整种子液含量至OD600为1.0,分别以0.5%的接种量转接WT和ΔompR至50 mL的LB液体培养基中,分别于27 ℃和37 ℃,180 r·min−1培养,每隔3 h取样测定菌液的OD600值,绘制生长曲线。
1.2.5 产灵菌红素能力的检测
用接种环分别挑取野生型菌株WT,转座子突变菌株FZSF02 ompR:: Tn5和基因敲除菌株FZSF02ΔompR划线于分为三区的LB固体琼脂平板上,平板倒置于27℃的恒温培养箱中培养36 h后观察菌株的产色能力。
1.2.6 qPCR试验
用细菌RNA提取试剂盒(TIANGEN,DP430)提取菌株的总RNA,应用FastKing gDNA Dispelling RT SuperMix(TIANGEN,KR118)进行反转录获取菌株cDNA,应用TransStart® Green qPCR SuperMix (TransGen,AQ101)进行qPCR扩增。qPCR以16SrDNA作为内参基因(引物为16SF和16SR),分别应用引物pigAF/pigAR,pigFF/pigFR和pigNF/pigNR检测灵菌红素合成基因簇中pigA,pigF和pigN基因的转录水平变化。qPCR引物序列信息见表1。
1.2.7 细菌生物被膜形成能力测定
分别挑取WT和ompR菌株单菌落至50 mL液体培养基中,于37 ℃、180 r·min−1培养过夜作为种子液,调整2个种子液含量至OD600值为1.0后以0.5%的接种量转接至50 mL的LB液体培养基中,从接种后的培养基中吸取200 μL至96孔细胞培养板中分别置于27 ℃和37 ℃培养,设置多个复孔作为重复。培养36 h测定各孔OD600后将培养板中的菌液吸出,用PBS缓冲液清洗各孔5次,倒置在滤纸上10 min以彻底除去孔中的PBS,向各孔加入200 μL 1%的结晶紫染料,染色30 min后吸出结晶紫,用PBS清洗各孔5次,倒置放置培养板10 min除去残留的PBS,用200 μL无水乙醇溶解各孔中的结晶紫,测定各孔OD595值。菌株的产膜能力用各菌株OD595/OD600值表示。
1.2.8 细菌运动性测定
配制琼脂含量分别为0.3%(m/V)和0.7%(m/V)的LB琼脂平板,分别吸取2 μL OD600值为2.0的WT和ΔompR菌液滴至各平板的中心位置,置于27℃培养箱培养,观察琼脂含量为0.3%(m/V)的平板上菌株的游动(swimming)能力,0.7%琼脂平板上菌株的涌动(swarming)能力。
1.2.9 细菌产酶能力测定
制备脱脂奶粉含量为1%(m/V)的LB琼脂平板用以检测蛋白酶产生能力,吐温80含量为1%(V/V)的LB琼脂平板用以检测脂肪酶产生能力,应用血琼脂平板检测菌株的溶血素产生能力。将2 μL OD600值为1.0的菌液滴于琼脂平板上,置于27 ℃培养48 h通过观察蛋白酶和脂肪酶酶活圈的有无和大小比较菌株的产酶能力;27 ℃培养24 h和7 d观察溶血素活性。
1.2.10 菌株胁迫耐受性测定
菌株的胁迫耐受性实验参考Brzostek等[20]和Gao等[21]的方法进行。菌株分别在pH 3.0处理10 min、55 ℃处理5 min、15 mmol·L−1过氧化氢处理10 min和2 mol·L−1的氯化钠处理1 h后稀释涂布LB琼脂平板进行菌落计数,以不经胁迫处理的菌株的菌数作为对照,分别计算经各胁迫因素处理后菌株的存活率。
2. 结果与分析
2.1 OmpR序列分析
粘质沙雷氏菌FZSF02菌株ompR基因有720个碱基(NCBI序列号:QJU42212.1),编码由239个氨基酸组成的OmpR蛋白。序列比对分析显示其氨基酸序列在不同物种间较为保守,与从前100个目标序列(Max target sequences)中所选代表不同属种的20株菌的相似性为99.16%~100.00%,系统发育树也显示出类似结果(图1)。其中,与3株沙雷氏菌属菌株的序列相似性为100.00%,与大肠杆菌等其他序列相似性为99.16%~99.58%。
2.2 基因敲除菌株的构建及鉴定
应用同源重组法构建ompR缺失菌株,挑取具卡那霉素抗性的疑似单菌落应用引物OmpRFF和OmpRBR验证,分别以FZSF02野生株和Tn5转座子突变株FZSF02 ompR:: Tn5作为对照。结果(图2)显示,以野生株FZSF02基因组为模板扩增得到大小为720 bp的单一条带,为ompR完整序列;以FZSF02 ompR:: Tn5基因组为模板扩增得到1 920 bp的单一条带,该序列由1 200 bp的Tn5转座子序列和被其插入的720 bp的ompR序列组成;以FZSF02ΔompR基因组为模板扩增得到大小为1 540 bp的条带,该序列由ompR上游300 bp,下游300 bp和中间940 bp的卡那抗性基因组成。琼脂糖凝胶电泳和测序结果均表明由卡那抗性基因替换掉120 bp ompR基因序列的ompR缺失菌株FZSF02ΔompR构建成功。
![]() 图 2 粘质沙雷氏菌FZSF02菌株ompR基因敲除验证注:应用引物OmpRFF和OmpRBR进行PCR。泳道M:DNA分子量标准;泳道1:FZSF02野生型;泳道2:ompR基因Tn5转座子插入突变体FZSF02 ompR:: Tn5;泳道3:ompR基因被敲除120 bp的菌株FZSF02ΔompR。Figure 2. Agarose gel electrophoresis identification of ompR-knockout S. marcescens FZSF02Note:PCR were carried out with primers OmpRFF and OmpRBR. Lane M: DNA marker; Lane 1: wild type FZSF02; Lane 2: FZSF02 ompR:: Tn5 with ompR inserted by Tn5 transposon; Lane 3: FZSF02ΔompR with 120 bp ompR-knockout.
图 2 粘质沙雷氏菌FZSF02菌株ompR基因敲除验证注:应用引物OmpRFF和OmpRBR进行PCR。泳道M:DNA分子量标准;泳道1:FZSF02野生型;泳道2:ompR基因Tn5转座子插入突变体FZSF02 ompR:: Tn5;泳道3:ompR基因被敲除120 bp的菌株FZSF02ΔompR。Figure 2. Agarose gel electrophoresis identification of ompR-knockout S. marcescens FZSF02Note:PCR were carried out with primers OmpRFF and OmpRBR. Lane M: DNA marker; Lane 1: wild type FZSF02; Lane 2: FZSF02 ompR:: Tn5 with ompR inserted by Tn5 transposon; Lane 3: FZSF02ΔompR with 120 bp ompR-knockout.2.3 生长能力检测
生长曲线显示FZSF02和FZSF02ΔompR在27 ℃(图3-A)和37℃(图3-B)条件下在LB液体培养基中的生长能力相似。FZSF02在27 ℃培养条件下24 h OD600值达到最大值6.60,48 h降至6.13;菌株ΔompR 24 h最高值为6.44,48 h降至6.27。37 ℃培养条件下,FZSF02在第21 h OD600值达到最大5.74,此后逐步降低至48 h 4.1;ΔompR 21 h达到最大值5.80,48 h降低至4.0。上述数据说明ompR基因的缺失对该菌的生长无明显影响。
2.4 OmpR缺失对FZSF02产灵菌红素的影响
LB固体平板试验显示,ompR 转座子插入突变菌株FZSF02 ompR::Tn5和缺失菌株FZSF02ΔompR均丧失灵菌红素产生能力(图4-A)。Pig基因簇大小约20 kb,由14个基因pigA-pigN按顺序排列;pigA,pigF和pigN分别是基因簇第1个、第6个和第14个基因,上述三个基因的表达水平可以反映该基因簇总体表达水平。 qPCR显示,FZSF02ΔompR菌株中灵菌红素合成基因簇中pigA,pigF和pigN基因的表达水平分别降低为野生型菌株的3.8%,2.0%和2.1%(图4-B),说明OmpR对灵菌红素合成的影响是通过调控灵菌红素合成基因簇的转录实现的。
![]() 图 4 OmpR对Serratia marcescens FZSF02产灵菌红素能力及灵菌红素合成基因表达的影响注:A:野生型菌株FZSF02, ompR 转座子突变菌株和ompR敲除菌株在LB琼脂平板上的生长情况。FZSF02, FZSF02 ompR:: Tn5 and FZSF02ΔompR分别代表野生型菌株,Tn5插入突变菌株和ompR敲除菌株。B:ompR基因敲除对零菌红素合成基因的影响。pigA, pigF和pigN为零菌红素合成基因簇上的3个基因。Log2倍数变化值代表上述三个基因在基因敲除菌株FZSF02ΔompR中相对野生型菌株的表达量变化。Figure 4. Effects of OmpR on prodigiosin-producing ability and expressions of prodigiosin biosynthesis genes of S. marcescens FZSF02Note:A: Prodigiosin-producing abilities of FZSF02, ompR mutant strain, and ompR-knockout strain on LB agar at 27 ℃. FZSF02, FZSF02 ompR:: Tn5, and FZSF02ΔompR were WT strain, Tn5 transposon insertion strain, and ompR-knockout strain, respectively. B: Deletion of ompR on expression of prodigiosin biosynthesis genes. pigA, pigF, and pigN were 3 genes in prodigiosin biosynthesis gene cluster. Log 2-fold change values represent expression levels of these genes in FZSF02ΔompR as compared to WT FZSF02.
图 4 OmpR对Serratia marcescens FZSF02产灵菌红素能力及灵菌红素合成基因表达的影响注:A:野生型菌株FZSF02, ompR 转座子突变菌株和ompR敲除菌株在LB琼脂平板上的生长情况。FZSF02, FZSF02 ompR:: Tn5 and FZSF02ΔompR分别代表野生型菌株,Tn5插入突变菌株和ompR敲除菌株。B:ompR基因敲除对零菌红素合成基因的影响。pigA, pigF和pigN为零菌红素合成基因簇上的3个基因。Log2倍数变化值代表上述三个基因在基因敲除菌株FZSF02ΔompR中相对野生型菌株的表达量变化。Figure 4. Effects of OmpR on prodigiosin-producing ability and expressions of prodigiosin biosynthesis genes of S. marcescens FZSF02Note:A: Prodigiosin-producing abilities of FZSF02, ompR mutant strain, and ompR-knockout strain on LB agar at 27 ℃. FZSF02, FZSF02 ompR:: Tn5, and FZSF02ΔompR were WT strain, Tn5 transposon insertion strain, and ompR-knockout strain, respectively. B: Deletion of ompR on expression of prodigiosin biosynthesis genes. pigA, pigF, and pigN were 3 genes in prodigiosin biosynthesis gene cluster. Log 2-fold change values represent expression levels of these genes in FZSF02ΔompR as compared to WT FZSF02.2.5 ompR缺失影响FZSF02生物被膜形成能力
生物被膜测定结果显示ompR部分缺失后生物被膜形成能力降低,37 ℃培养条件下FZSF02ΔompR的生物膜量较FZSF02野生型菌株降低37.5%(P<0.01),27 ℃培养条件下生物被膜量降低15.1%(P<0.01),表明OmpR参与了该菌生物被膜的合成(图5)。
2.6 OmpR对FZSF02运动和产酶能力的影响
检测野生型菌株和基因敲除菌株FZSF02ΔompR在琼脂含量为0.3%的LB平板上的游动能力(swimming),菌株FZSF02和ΔompR的菌落直径分别为7.32 cm和7.18 cm;在0.7%琼脂含量的LB平板上的涌动能力(swarming)试验显示FZSF02和ΔompR的菌落直径分别为5.51 cm和5.67 cm,数值无显著性差异(图6),说明OmpR对该菌的运动性无明显影响。野生型菌株WT和FZSF02ΔompR均有产蛋白酶能力(酶活圈直径分别为2.1 cm和2.2 cm)(图7-A)和脂肪酶能力(酶活圈直径分别为1.6 cm和1.7 cm)(图7-B),从酶活圈直径评价二者产酶能力无明显区别。WT和FZSF02ΔompR溶血能力相似,在培养第24 h时均无明显溶血圈(图7-C),培养至第7天时都产生微弱的溶血圈(图7-D)。
![]() 图 6 Serratia marcescens FZSF02和FZSF02ΔompR的运动能力注:在0.3%(w/v)和0.7%(w/v)琼脂的LB固体平板上检测Serratia marcescens FZSF02和基因敲除菌株FZSF02ΔompR运动的菌落直径。Figure 6. Mobility of S. marcescens FZSF02 and FZSF02ΔompRNote:Mobility diameters of S. marcescens FZSF02 and FZSF02ΔompR were assayed on solid LB plates containing 0.3% (w/v) and 0.7% (w/v) agar, respectively.
图 6 Serratia marcescens FZSF02和FZSF02ΔompR的运动能力注:在0.3%(w/v)和0.7%(w/v)琼脂的LB固体平板上检测Serratia marcescens FZSF02和基因敲除菌株FZSF02ΔompR运动的菌落直径。Figure 6. Mobility of S. marcescens FZSF02 and FZSF02ΔompRNote:Mobility diameters of S. marcescens FZSF02 and FZSF02ΔompR were assayed on solid LB plates containing 0.3% (w/v) and 0.7% (w/v) agar, respectively.![]() 图 7 Serratia marcescens FZSF02和FZSF02ΔompR的产酶能力注:WT和ΔompR分别代表Serratia marcescens FZSF02野生型菌株和ompR敲除菌株。A:在含1% (w/v)脱脂奶粉的LB固体琼脂培养基上的蛋白酶产生能力。B:在含1%(v/v) tween 80的固体琼脂培养基上的脂肪酶产生能力。C和D:在血琼脂培养基上27 ℃条件下培养24小时和7天观察溶血素产生能力。Figure 7. Enzyme-producing abilities of S. marcescens FZSF02 and FZSF02ΔompRNote:WT andΔompR represent WT and ompR-knockout S. marcescens FZSF02, respectively. A: Protease-producing ability on LB agar plates containing 1% (w/v) of skim milk. B: Lipase-producing ability on LB agar plates containing 1% (v/v) of tween 80. C and D: Hemolysin-producing abilities on blood agar base medium after incubation at 27 ℃ for 24 h (C) and 7 days (D), respectively.
图 7 Serratia marcescens FZSF02和FZSF02ΔompR的产酶能力注:WT和ΔompR分别代表Serratia marcescens FZSF02野生型菌株和ompR敲除菌株。A:在含1% (w/v)脱脂奶粉的LB固体琼脂培养基上的蛋白酶产生能力。B:在含1%(v/v) tween 80的固体琼脂培养基上的脂肪酶产生能力。C和D:在血琼脂培养基上27 ℃条件下培养24小时和7天观察溶血素产生能力。Figure 7. Enzyme-producing abilities of S. marcescens FZSF02 and FZSF02ΔompRNote:WT andΔompR represent WT and ompR-knockout S. marcescens FZSF02, respectively. A: Protease-producing ability on LB agar plates containing 1% (w/v) of skim milk. B: Lipase-producing ability on LB agar plates containing 1% (v/v) of tween 80. C and D: Hemolysin-producing abilities on blood agar base medium after incubation at 27 ℃ for 24 h (C) and 7 days (D), respectively.2.7 ompR对FZSF02响应胁迫的影响
检测了野生型菌株和基因敲除菌株FZSF02ΔompR在高温,低pH,高盐和强氧化环境下的存活能力。55 ℃高温处理后野生型菌株和∆ompR的存活率分别为0.15%和0.11%;pH3.0条件下处理后存活率分别为22.72%和28.35%;2 mol·L−1 NaCl处理后的存活率分别为92.45%和90.65%;氧化剂0.22 mol·L−1 H2O2处理后的存活率分别为22.40%和23.84%。说明上述胁迫条件下二者存活率无明显差异(表2)。
表 2 胁迫条件下WT与∆ompR的存活率Table 2. Survival rates of WT and ∆ompR under stress (单位:%)胁迫条件
Stress factorWT 存活率
Survival rate of WT∆ompR 存活率
Survival rate of ∆ompRpH=3 22.72±3.40 a 28.35±4.10 a 55 ℃ 0.15±0.01 a 0.11±0.05 a 2 mol·L−1 NaCl 92.45±2.60 a 90.65±3.80 a 0.22 mol·L−1 H2O2 22.40±1.30 a 23.84±1.10 a 3. 讨论与结论
本研究成功构建了粘质沙雷氏菌FZSF02的ompR敲除菌株,通过对突变体和野生型的对比研究了OmpR对粘质沙雷氏菌FZSF02灵菌红素产生能力,生物膜形成能力,产酶能力,运动性和适应胁迫等生物学特性的影响。
粘质沙雷氏菌合成灵菌红素受已报道的30多种基因调控 [22],双组分调控系统是其中重要的一类调控基因。目前在粘质沙雷氏菌菌株中发现的参与色素合成调控的双组分系统有PigQ/PigW[13](与BarA/UvrY系统高度同源 [23])、PhoB/PhoR [14]、RssB/RssA[16]和 EepR/EepS [15]。PigQ/PigW,PhoB/PhoR和eepR/eepS双组分系统是通过效应蛋白直接与pigA上游启动子区结合激活灵菌红素基因簇的表达而正向调控灵菌红素合成[13-15];RssB/RssA调控系统通过RssB与pigA上游启动子直接结合抑制灵菌红素基因簇表达从而负调控灵菌红素的合成 [16]。本研究EnvZ/OmpR双组分调控系统效应蛋白基因ompR敲除后菌株完全丧失灵菌红素合成能力,对pigA-N基因簇中分别处于基因簇上游、中游和下游的pigA、pigF和pigN基因转录水平应用qPCR检测,结果显示ompR敲除菌株中灵菌红素合成基因簇中3个基因转录水平明显降低,说明转录调控因子OmpR正调控该菌株灵菌红素的合成,该调控可能通过影响整个基因簇的转录实现。ompR是本研究新鉴定的参与粘质沙雷氏菌灵菌红素合成调控的双组分系统效应基因,有别于已报道的其他4种双组分调控系统,其是否与PigQ/PigW,PhoB/PhoR和eepR/eepS双组份系统类似通过与pigA上游启动子直接结合从而正调控灵菌红素基因簇的表达有待通过凝胶迁移试验和DNaseI足迹试验等进一步研究。
OmpR对多种微生物生物膜的形成有重要影响。ompR缺失或突变降低大肠杆菌[24-25],Yersinia enterocolitica [26]和肠炎沙门氏菌Salmonella enteritidis[27]生物被膜的形成,但是对Acinetobacter baumannii Strain AB5075生物被膜形成能力无影响 [28]。菌株FZSF02 ompR缺失菌株生物被膜形成量在37 ℃时降低37.5%,降低程度与Yersinia enterocolitica的ompR突变株类似(降低33%)[26],小于肠炎沙门氏菌Salmonella enteritidis(降低75%以上)[27]。上述结果暗示OmpR对细菌生物被膜形成的影响有一定的普遍性。
OmpR影响细菌多个微生物学特性,虽然在不同种属间OmpR序列高度保守,但是在不同菌株中其作用不同甚至相反。ompR敲除后Acinetobacter baumannii Strain AB5075的游动和涌动能力下降[26],但是在Xenorhabdus nematophila[29]中运动能力显著增强。Xenorhabdus nematophila ompR基因敲除菌株蛋白酶,脂肪酶和溶血能力均明显增强[29];Yersinia enterocolitica菌株ompR基因敲除后对高盐渗透压、低pH、过氧化氢和高温胁迫条件的适应性均明显降低[20];但是Yersinia pestis ompR基因敲除菌株对高盐、低pH和高温条件较野生型菌株更为敏感,但是对过氧化氢耐受性增强[21]。本研究对菌株FZSF02上述生物学特性进行探讨,显示ompR敲除后该菌运动性,产酶能力及环境胁迫的适应性均无明显变化。说明OmpR在粘质沙雷氏菌FZSF02菌株中功能与已报道的菌株不同,这反映了OmpR在不同微生物中调控功能的多样性和复杂性。
综上,本研究发现在粘质沙雷氏菌FZSF02菌株中OmpR特异性正调控灵菌红素的合成,同时一定程度上影响生物被膜的形成。EnvZ/OmpR代表一个新的灵菌红素合成调控系统,其具体调控机制有待深入研究。
-
表 1 17份南安石亭群体种茶树种质资源
Table 1 Germplasm resources of 17 C.sinensis plants planted in Shiting population, Nan'an
编号
Code茶树种质
Germplasm resour测s长宽比
Aspect ratio叶面积
Leaf area/cm2叶形
Blade shape叶色
Leaf color1 ST1 2.87 10.63 长椭圆形 黄绿色 2 ST2 2.83 10.47 长椭圆形 黄绿色 3 ST3 2.54 10.25 长椭圆形 黄绿色 4 ST4 2.32 10.15 椭圆形 黄绿色 5 ST5 2.61 14.31 长椭圆形 黄绿色 6 ST6 3.10 9.56 披针形 黄绿色 7 ST7 2.82 9.55 长椭圆形 深绿色 8 ST8 2.20 6.16 椭圆形 黄绿色 9 ST9 2.25 16.13 椭圆形 深绿色 10 ST10 2.37 5.99 椭圆形 深绿色 11 ST11 2.35 6.58 椭圆形 黄绿色 12 ST12 2.18 17.61 椭圆形 黄绿色 13 ST13 2.44 10.68 椭圆形 深绿色 14 ST14 2.52 9.34 长椭圆形 黄绿色 15 ST15 2.26 8.37 椭圆形 深绿色 16 ST16 2.17 13.65 椭圆形 黄绿色 17 ST17 2.50 11.83 椭圆形 黄绿色 表 2 南安石亭群体种茶树种质资源儿茶素组分含量
Table 2 Catechins of Shiting plantation tea germplasms
(单位:mg·g−1) 儿茶素类
CatechinsST2 ST3 ST5 ST6 ST8 ST10 ST11 ST12 ST14 ST17 非酯
型儿
茶素
Non-ester
catechinsGC 0.35±0.02f 0.96±0.05a 0.42±0.01ef 0.48±0.02def 0.85±0.02ab 0.49±0.06def 0.76±0.03bc 0.61±0.12cde 0.88±0.02ab 0.65±0.30cd EGC 23.68±1.41f 28.91±2.25e 38.57±2.38c 24.79±2.07ef 56.41±3.73a 29.40±3.30d 38.44±3.66c 45.12±3.11b 44.39±1.16b 31.80±0.40d C 1.72±0.06 2.15±0.18a 1.93±0.09ab 1.98±0.25ab 1.95±0.30ab 1.36±0.31 c 1.78±0.27ab 0.83±0.09de 0.91±0.08d 0.51±0.01e EC 13.83±1.40cd 11.89±0.79e 15.06±0.36c 7.06±0.49f 18.72±0.55a 8.13±0.34f 13.29±0.88d 17.44±1.07b 10.96±0.47e 7.82±0.13f 总量 39.58±2.06de 43.91±3.34d 55.98±2.77c 34.31±2.88e 77.93±4.35a 39.38±3.83de 54.27±4.77c 64.00±4.44b 57.14±1.74c 40.78±0.38d 酯型儿
茶素
Ester
catechinsEGCG 92.51±6.53a 52.21±3.22d 57.99±1.27cd 98.30±6.29a 61.37±2.20c 76.02±4.76b 40.30±3.97e 57.12±5.35cd 63.94±2.28c 39.60±0.91e EGCG3" Me 12.97±0.88b 13.27±0.66b 1.66±0.11e — 7.11±0.18c — 6.02±0.52d 6.55±0.48cd 6.36±0.25cd 18.18±0.14a ECG 31.75±2.02a 14.82±0.58e 17.63±0.43c 19.92±0.96b 17.16±0.47cd 14.93±0.70e 11.18±0.85f 15.77±1.11de 11.64±0.38f 7.58±0.12g CG 0.66±0.03c 0.94±0.05b 0.65±0.03c 0.62±0.02c — 0.65±0.02c — — — 1.41±0.08a 总量 137.89±9.45a 81.24±4.49d 77.93±1.79d 118.84±7.25b 85.64±2.86cd 91.6±5.44c 57.50±5.35f 79.44±6.88d 81.94±2.89d 66.77±0.88e 总儿茶素
Total
catechins177.47±11.15a 125.15±7.81ef 133.91±4.28de 153.15±9.93bc 163.57±6.84ab 130.98±7.64de 111.77±10.00fg 143.44±11.29cd 139.08±4.56cde 107.55±1.02g 儿茶素
苦涩味指数
Catechins
bitterness and
astringency index9.54±1.07c 6.90±0.13d 6.74±0.07de 15.87±0.07a 6.57±0.16de 12.73±0.30b 6.02±0.16e 6.49±0.15de 10.18±0.15c 9.56±0.18c 儿茶素
品质指数
Catechin
quality index524.60±7.00a 232.12±5.47d 196.46±10.04e 477.71±17.2b 139.46±6.84gh 311.54±33.00c 133.99±3.89h 161.41±3.74fg 170.25±2.58f 148.39±4.63fgh 数值表示3个样品均值±标准差;同行不同小写字母表示差异显著(P < 0.05);“—”表示未检测出。表3 同。
Data are standard deviation ± mean of triplicated samples; those with different lowercase letters on same row indicate significant differences at P<0.05; "—" indicates not detected. Same for Table 3.表 3 南安石亭群体种茶树种质资源氨基酸的组分含量
Table 3 Amino acids of Shiting plantation tea germplasms
(单位:mg·g−1) 氨基酸组分
Amino acid
componentsST2 ST3 ST5 ST6 ST8 ST10 ST11 ST12 ST14 ST17 鲜味类
FlavorThea 3.27±0.14e 9.98±0.13a 5.54±0.17d 2.07±0.12g 1.61±0.28h 7.17±0.38b 6.50±0.36c 2.62±0.19f 2.37±0.06fg 2.10±0.00g Pro 0.29±0.01e 0.69±0.02a 0.31±0.01e 0.52±0.04bc 0.37±0.18de 0.36±0.02de 0.47±0.04cd 0.60±0.02ab 0.40±0.04de 0.38±0.03de Glu 2.27±0.15bc 2.53±0.29b 0.61±0.05f 1.13±0.04e 0.97±0.14ef 1.93±0.00cd 1.76±0.23d 3.90±0.65a 4.09±0.13a 0.14±0.02g GABA 0.06±0.00a 0.06±0.00a 0.06±0.00a 0.06±0.00a 0.06±0.00a 0.06±0.00a 0.07±0.00a 0.07±0.00a 0.07±0.00a 0.07±0.00a Asp 0.30±0.14cd 0.59±0.01b 0.24±0.03de 0.88±0.04a 0.15±0.02ef 0.31±0.08cd 0.17±0.04ef 0.34±0.05cd 0.39±0.07c 0.12±0.01f 总量 6.20±0.26f 13.85±0.24a 6.76±0.19e 4.66±0.13g 3.16±0.35g 9.83±0.29b 8.96±0.2c 7.54±0.49d 7.31±0.09d 0.81±0.01h 甜味类
SweetnessAla 0.13±0.04f 0.28±0.01de 0.14±0.03f 0.18±0.04ef 0.46±0.1c 0.47±0.06c 0.93±0.09b 0.39±0.02cd 0.16±0.04ef 2.00±0.17a Thr 0.22±0.03e 0.53±0.03a 0.27±0.02d 0.41±0.03b 0.17±0.04e 0.35±0.02c 0.36±0.03c 0.36±0.02c 0.35±0.04c 0.21±0.02e Asn 0.28±0.03cd 0.57±0.2a 0.24±0.07cd 0.6±0.01a 0.19±0.04d 0.37±0.12bc 0.29±0.04cd 0.35±0.02bcd 0.48±0.06ab 0.24±0.02cd Ser 0.37±0.01f 1.24±0.06a 0.47±0.05ef 0.8±0.08c 0.55±0.03def 1.00±0.08b 0.66±0.31cde 0.8±0.02c 0.75±0.05cd 0.43±0.07f Gly 0.04±0.01d 0.06±0.02cd 0.25±0.03a 0.09±0.02cd 0.12±0.07c 0.06±0.04cd 0.19±0.01b 0.05±0.03d 0.06±0.05cd 0.1±0.04cd Gln 3.15±0.27b 3.10±0.02b 0.47±0.10f 1.59±0.08e 1.00±0.17e 3.76±0.18a 2.58±0.34c 2.99±0.27b 3.69±0.13a 0.18±0.03f 总量 4.22±0.13d 5.80±0.05ab 1.85±0.03g 3.96±0.22d 2.45±0.28f 5.94±0.37a 4.88±0.17c 4.93±0.3c 5.40±0.35b 3.03±0.19e 芳香类
AromaticLys 0.21±0.04cd 0.62±0.03a 0.15±0.02de 0.42±0.03b 0.19±0.08cd 0.24±0.02c 0.25±0.01c 0.38±0.02b 0.23±0.03c 0.11±0.04e Tyr 0.53±0.05cd 0.61±0.03c 0.49±0.04de 0.81±0.07b 0.54±0.1cd 0.41±0.04ef 0.85±0.07b 2.24±0.03a 0.55±0.01cd 0.36±0.08g Trp 0.47±0.04a 0.27±0.02c 0.16±0.04e 0.33±0.03b 0.17±0.04e 0.16±0.00e 0.17±0.01e 0.28±0.02c 0.22±0.01d 0.08±0.02f 总量 1.21±0.13c 1.5±0.02b 0.79±0.03e 1.56±0.04b 0.90±0.16de 0.81±0.06e 1.27±0.08c 2.9±0.05a 1.00±0.02d 0.55±0.11f 苦味类
BittersArg 2.81±0.15d 8.25±0.46a 4.93±0.13c 1.86±0.12e 0.98±0.36f 5.91±0.27b 6.30±0.31b 2.18±0.3e 2.11±0.25e 0.14±0.01g Leu 0.28±0.01ef 1.07±0.02a 0.45±0.06bc 0.5±0.04b 0.33±0.04de 0.27±0.02ef 0.24±0.03fg 0.39±0.06cd 0.34±0.02de 0.2±0.04g Ile 0.31±0.02cd 0.91±0.21a 0.45±0.09bc 0.48±0.03b 0.06±0.03f 0.22±0.00def 0.22±0.05def 0.29±0.11cde 0.38±0.09bcd 0.14±0.03ef His 0.11±0.02cd 0.15±0.02b 0.09±0.01d 0.19±0.00a 0.11±0.04cd 0.11±0.02cd 0.12±0.02bcd 0.14±0.01bc 0.11±0.02bcd 0.09±0.02d Val 0.18±0.03e 0.49±0.02a 0.12±0.01f 0.40±0.01b 0.06±0.03g 0.22±0.03d 0.21±0.02de 0.33±0.02c 0.23±0.01d 0.06±0.02g 总量 3.68±0.18d 10.86±0.36a 6.04±0.29c 3.43±0.15de 1.54±0.38f 6.73±0.22b 7.09±0.29b 3.33±0.27de 3.17±0.31e 0.62±0.08g 其他
Otherβ-ABA 0.09±0.01e 0.11±0.01e 2.42±0.27b 0.08±0.01e 0.85±0.26d 0.16±0.04e 2.67±0.22a 1.24±0.13c 0.07±0.01e 0.90±0.04d sar 0.17±0.01c 0.22±0.04bc 0.23±0.00bc 0.31±0.13abc 0.19±0.03bc 0.34±0.14ab 0.33±0.09abc 0.28±0.06abc 0.41±0.06a 0.32±0.10abc 总氨基酸 15.55±0.39f 32.32±0.44a 18.09±0.7e 13.70±0.08g 9.15±0.68h 24.87±0.57c 25.32±0.19b 20.21±0.52d 17.47±0.8e 8.35±0.27i -
[1] 阚能才. 茶树起源与川渝野生茶树分布研究 [J]. 西南农业学报, 2013, 26(1):382−385. DOI: 10.3969/j.issn.1001-4829.2013.01.080 KAN N C. Study on geographical distribution of wild tea trees in Sichuan and Chongqing, and origin of tea tree [J]. Southwest China Journal of Agricultural Sciences, 2013, 26(1): 382−385. (in Chinese) DOI: 10.3969/j.issn.1001-4829.2013.01.080
[2] 黄天柱, 廖渊泉. 石亭茶香飘海宇 [J]. 农业考古, 1991(4):227. HUANG T Z, LIAO Y Q. Shi Ting Cha Xiang Piao Hai Yu [J]. Agricultural Archaeology, 1991(4): 227. (in Chinese)
[3] 林金良, 陈育才. 九日山和石亭绿茶文化资源探讨[J]. 福建茶叶, 2020, 42(11): 307-309. LIN J L, CHEN Y C. Discussion on jiuri mountain heshiting green tea cultural resources[J]. Tea in Fujian, 2020, 42(11): 307-309. (in Chinese)
[4] WANG B Y, TAN H W, FANG W P, et al. Developing single nucleotide polymorphism (SNP) markers from transcriptome sequences for identification of Longan (Dimocarpus longan) germplasm [J]. Horticulture Research, 2015, 2: 14065. DOI: 10.1038/hortres.2014.65
[5] XU C, REN Y H, JIAN Y Q, et al. Development of a maize 55 K SNP array with improved genome coverage for molecular breeding [J]. Molecular Breeding, 2017, 37(3): 20. DOI: 10.1007/s11032-017-0622-z
[6] 郭灿, 皮发娟, 吴昌敏, 等. 基于GBS测序的全基因组SNP揭示贵州地方茶组植物资源的亲缘关系 [J]. 南方农业学报, 2021, 52(3):660−670. DOI: 10.3969/j.issn.2095-1191.2021.03.014 GUO C, PI F J, WU C M, et al. Genome-wide SNP developed by genotyping-by-sequencing revealed the phylogenetic relationship of Sect. Thea(L. ) Dyer resources in Guizhou [J]. Journal of Southern Agriculture, 2021, 52(3): 660−670. (in Chinese) DOI: 10.3969/j.issn.2095-1191.2021.03.014
[7] LIN Y, YU W T, CAI C P, et al. Rapid varietal authentication of oolong tea products by microfluidic-based SNP genotyping [J]. Food Research International, 2022, 162: 111970. DOI: 10.1016/j.foodres.2022.111970
[8] LIU C G, YU W T, CAI C P, et al. Genetic diversity of tea plant (Camellia sinensis (L.) kuntze) germplasm resources in Wuyi mountain of China based on single nucleotide polymorphism (SNP) markers [J]. Horticulturae, 2022, 8(10): 932. DOI: 10.3390/horticulturae8100932
[9] CHEN Y J, NIU S Z, DENG X Y, et al. Genome-wide association study of leaf-related traits in tea plant in Guizhou based on genotyping-by-sequencing [J]. BMC Plant Biology, 2023, 23(1): 196. DOI: 10.1186/s12870-023-04192-0
[10] LIAO Y Y, ZHOU X C, ZENG L T. How does tea (Camellia sinensis) produce specialized metabolites which determine its unique quality and function: A review [J]. Critical Reviews in Food Science and Nutrition, 2022, 62(14): 3751−3767. DOI: 10.1080/10408398.2020.1868970
[11] WILLIAMS J L, EVERETT J M, D’CUNHA N M, et al. The effects of green tea amino acid L-theanine consumption on the ability to manage stress and anxiety levels: A systematic review [J]. Plant Foods for Human Nutrition, 2020, 75(1): 12−23. DOI: 10.1007/s11130-019-00771-5
[12] 江新凤, 李琛, 石旭平, 等. 高效液相色谱法对“黄金菊” 茶中儿茶素和氨基酸组分含量的测定 [J]. 食品研究与开发, 2021, 42(5):172−176. DOI: 10.12161/j.issn.1005-6521.2021.05.029 JIANG X F, LI C, SHI X P, et al. HPLC determination of catechin and amino acid components in Camellia sinensis ‘huangjinju’ [J]. Food Research and Development, 2021, 42(5): 172−176. (in Chinese) DOI: 10.12161/j.issn.1005-6521.2021.05.029
[13] UNNO K, NAKAMURA Y. Green tea suppresses brain aging [J]. Molecules, 2021, 26(16): 4897. DOI: 10.3390/molecules26164897
[14] JIN J Q, JIANG C K, YAO M Z, et al. Baiyacha, a wild tea plant naturally occurring high contents of theacrine and 3″-methyl-epigallocatechin gallate from Fujian, China [J]. Scientific Reports, 2020, 10(1): 9715. DOI: 10.1038/s41598-020-66808-x
[15] 金基强, 张晨禹, 马建强, 等. 茶树种质资源研究“十三五” 进展及“十四五” 发展方向 [J]. 中国茶叶, 2021, 43(9):42−49,76. DOI: 10.3969/j.issn.1000-3150.2021.09.006 JIN J Q, ZHANG C Y, MA J Q, et al. Research progress on tea germplasms during the 13th Five-Year Plan period and development direction in the 14th Five-Year Plan period [J]. China Tea, 2021, 43(9): 42−49,76. (in Chinese) DOI: 10.3969/j.issn.1000-3150.2021.09.006
[16] WANG P J, GU M Y, SHAO S X, et al. Changes in non-volatile and volatile metabolites associated with heterosis in tea plants (Camellia sinensis) [J]. Journal of Agricultural and Food Chemistry, 2022, 70(9): 3067−3078. DOI: 10.1021/acs.jafc.1c08248
[17] 江昌俊. 茶树育种学[M]. 北京: 中国农业出版社: 2005. [18] FANG W P, MEINHARDT L W, TAN H W, et al. Varietal identification of tea (Camellia sinensis) using nanofluidic array of single nucleotide polymorphism (SNP) markers [J]. Horticulture Research, 2014, 1: 14035. DOI: 10.1038/hortres.2014.35
[19] 樊晓静, 于文涛, 蔡春平, 等. 利用SNP标记构建茶树品种资源分子身份证 [J]. 中国农业科学, 2021, 54(8):1751−1772. DOI: 10.3864/j.issn.0578-1752.2021.08.014 FAN X J, YU W T, CAI C P, et al. Construction of molecular ID for tea cultivars by using of single-nucleotide polymorphism(SNP) markers [J]. Scientia Agricultura Sinica, 2021, 54(8): 1751−1772. (in Chinese) DOI: 10.3864/j.issn.0578-1752.2021.08.014
[20] 徐梦婷, 魏明秀, 陈潇敏, 等. 寿宁长叶1号等茶树新品系儿茶素和氨基酸组分分析 [J]. 茶叶学报, 2022, 63(1):20−26. DOI: 10.3969/j.issn.1007-4872.2022.01.004 XU M T, WEI M X, CHEN X M, et al. Catechins and amino acids in shouningchangye No. 1 and other new tea varieties [J]. Acta Tea Sinica, 2022, 63(1): 20−26. (in Chinese) DOI: 10.3969/j.issn.1007-4872.2022.01.004
[21] ZHANG Y N, YIN J F, CHEN J X, et al. Improving the sweet aftertaste of green tea infusion with tannase [J]. Food Chemistry, 2016, 192: 470−476. DOI: 10.1016/j.foodchem.2015.07.046
[22] 李芬, 陈春林, 田玉萍, 等. 云南不同品种大叶种茶树生化成分季节变化特征分析 [J]. 食品与生物技术学报, 2022, 41(3):88−95. DOI: 10.3969/j.issn.1673-1689.2022.03.012 LI F, CHEN C L, TIAN Y P, et al. Seasonal Variation of Biochemical Components of Different Cultivars of Camellia sinensis var. assamica in Yunnan [J]. Journal of Food Science and Biotechnology, 2022, 41(3): 88−95. (in Chinese) DOI: 10.3969/j.issn.1673-1689.2022.03.012
[23] YU P H, HUANG H, ZHAO X, et al. Dynamic variation of amino acid content during black tea processing: A review [J]. Food Reviews International, 2023, 39(7): 3970−3983. DOI: 10.1080/87559129.2021.2015374
[24] HUANG X J, CAO H L, GUO Y L, et al. The dynamic change of oolong tea constitutes during enzymatic-catalysed process of manufacturing [J]. International Journal of Food Science & Technology, 2020, 55(12): 3604−3612.
[25] LIU Z Y, RAN Q S, LI Q, et al. Interaction between major catechins and umami amino acids in green tea based on electronic tongue technology [J]. Journal of Food Science, 2023, 88(6): 2339−2352. DOI: 10.1111/1750-3841.16543
[26] JIN J Q, MA J Q, MA C L, et al. Determination of catechin content in representative Chinese tea germplasms [J]. Journal of Agricultural and Food Chemistry, 2014, 62(39): 9436−9441. DOI: 10.1021/jf5024559
[27] 杨春, 吴昌敏, 石伟昌, 等. 黎平地方茶树资源生化成分多样性分析及优异单株鉴选 [J]. 种子, 2020, 39(10):63−66,72. YANG C, WU C M, SHI W C, et al. Diversity analysis of biochemical components of local tea tree resources in Liping Region and excellent individual plant selection [J]. Seed, 2020, 39(10): 63−66,72. (in Chinese)
[28] ZHANG M, ZHANG X, HO C T, et al. Chemistry and health effect of tea polyphenol (–)-epigallocatechin 3-O-(3-O-methyl)gallate [J]. Journal of Agricultural and Food Chemistry, 2019, 67(19): 5374−5378. DOI: 10.1021/acs.jafc.8b04837
[29] LI J H, CHEN S X, ZHU M Z, et al. Cluster analysis of the biochemical composition in 53 Sichuan EGCG3"Me tea resources [J]. IOP Conference Series: Materials Science and Engineering, 2017, 231: 012125. DOI: 10.1088/1757-899X/231/1/012125
[30] XU C H, LIANG L, LI Y H, et al. Studies of quality development and major chemical composition of green tea processed from tea with different shoot maturity [J]. LWT, 2021, 142: 111055. DOI: 10.1016/j.lwt.2021.111055
[31] JIANG H, YU F, QIN L, et al. Dynamic change in amino acids, catechins, alkaloids, and Gallic acid in six types of tea processed from the same batch of fresh tea (Camellia sinensis L. ) leaves [J]. Journal of Food Composition and Analysis, 2019, 77: 28−38. DOI: 10.1016/j.jfca.2019.01.005
[32] ZHANG L, CAO Q Q, GRANATO D, et al. Association between chemistry and taste of tea: A review [J]. Trends in Food Science & Technology, 2020, 101: 139−149.
[33] LI Y C, RAN W, HE C, et al. Effects of different tea tree varieties on the color, aroma, and taste of Chinese Enshi green tea [J]. Food Chemistry: X, 2022, 14: 100289. DOI: 10.1016/j.fochx.2022.100289
[34] HU S, HE C, LI Y C, et al. Changes of fungal community and non-volatile metabolites during pile-fermentation of dark green tea [J]. Food Research International, 2021, 147: 110472. DOI: 10.1016/j.foodres.2021.110472
-
期刊类型引用(1)
1. 丁霞飞,贾宪波,林陈强,庄振宏,陈济琛. 普城沙雷氏菌ACCC 02146产灵菌红素调控基因的鉴定. 福建农业学报. 2023(04): 485-496 .  本站查看
本站查看
其他类型引用(2)




 下载:
下载: